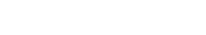チームリーダー
石井 佳誉
Ph.D.
先端NMR開発・応用研究チーム
[2025年3月 終了]
E-mail yoshitaka.ishii@riken.jp
当チームは理研横浜事業所において世界最高水準のNMR研究拠点を整備・発展してきました。この拠点をベースに世界を先導するNMRの方法論の開発や先端応用研究をおこないます。従来の手法では解析が困難なアミロイドタンパク質やRNA、ナノ生体材料等の生体分子の構造と分子レベルでの機能の解明を目指し、技術開発に取り組みます。更に、JSTのプロジェクト等で開発中の超高磁場NMR磁石に対応した新規NMR法の開発をおこない、従来の限界を超越したNMR技術基盤を確⽴するとともに、新研究領域や複合測定法を積極的に開拓します。
研究テーマ
- 構造生物化学
- 先端ナノ材料・高分子
- アルツハイマー病等の分子機構
主要論文
Ohyama T, Osawa T, Sekine SI, Ishii Y.
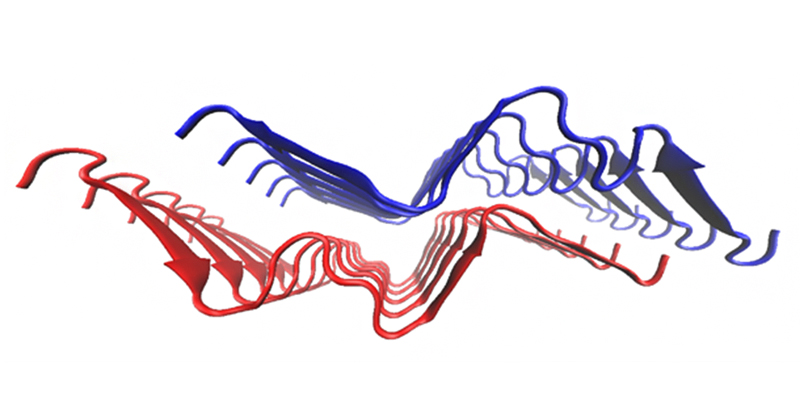
NMR Studies of Genomic RNA in 3' Untranslated Region Unveil Pseudoknot Structure that Initiates Viral RNA Replication in SARS-CoV-2.
JACS Au
4(4), 1323-1333 (2024)
doi: 10.1021/jacsau.3c00641
Sakuma K, Kobayashi N, Sugiki T, et al.
Design of complicated all-α protein structures.
Nature Structural & Molecular Biology
31(2), 275-282 (2024)
doi: 10.1038/s41594-023-01147-9
Tritrakarn T, Yamamoto K, Takahashi M, Okamura T.
New unifying metric for NMR/MRI probe evaluation based on optimized solenoid coil geometry.
Journal of Magnetic Resonance
358, 107602 (2024)
doi: 10.1016/j.jmr.2023.107602
Takatsu K, Kobayashi N, Wu N, et al.
Biophysical analysis of Gaussia luciferase bioluminescence mechanisms using a non-oxidizable coelenterazine.
BBA Advances
3, 100068 (2023)
doi: 10.1016/j.bbadva.2022.100068
Minami S, Kobayashi N, Sugiki T, et al.
Exploration of novel αβ-protein folds through de novo design.
Nature Structural & Molecular Biology
30(8), 1132-1140 (2023)
doi: 10.1038/s41594-023-01029-0
Kashihara K, Oouchi M, Kodama Y, et al.
High-Field Nuclear Magnetic Resonance Studies Reveal New Structural Landscape of Sulfur-Vulcanized Natural Rubber.
Biomacromolecules
23(11), 4481-4492 (2022)
doi: 10.1021/acs.biomac.2c00141
Matsunaga T, Okabe R, Ishii Y.
Efficient solvent suppression with adiabatic inversion for 1H-detected solid-state NMR.
Journal of Biomolecular NMR
(2021)
doi: 10.1007/s10858-021-00384-8
Wickramasinghe A, Xiao Y, Kobayashi N, et al.
Sensitivity-Enhanced Solid-State NMR Detection of Structural Differences and Unique Polymorphs in Pico- to Nanomolar Amounts of Brain-Derived and Synthetic 42-Residue Amyloid-β Fibrils.
Journal of the American Chemical Society
143(30), 11462-11472 (2021)
doi: 10.1021/jacs.1c03346
Matsunaga T, Matsuda I, Yamazaki T, Ishii Y.
Decoherence optimized tilted-angle cross polarization: A novel concept for sensitivity-enhanced solid-state NMR using ultra-fast magic angle spinning.
Journal of Magnetic Resonance
322, 106857 (2021)
doi: 10.1016/j.jmr.2020.106857
Koga R, Yamamoto M, Kosugi T, et al.
Robust folding of a de novo designed ideal protein even with most of the core mutated to valine.
Proceedings of the National Academy of Sciences of the United States of America
117(49), 31149-31156 (2020)
doi: 10.1073/pnas.2002120117
Ohyama T, Takahashi H, Sharma H, et al.
An NMR-based approach reveals the core structure of the functional domain of SINEUP lncRNAs.
Nucleic Acids Research
48(16), 9346-9360 (2020)
doi: 10.1093/nar/gkaa598
Xiao Y, Matsuda I, Inoue M, et al.
NMR-based site-resolved profiling of β-amyloid misfolding reveals structural transitions from pathologically relevant spherical oligomer to fibril.
the Journal of Biological Chemistry
295(2), 458-467 (2020)
doi: 10.1074/jbc.RA119.008522
Oouchi M, Ukawa J, Ishii Y, Maeda H.
Structural Analysis of the Terminal Groups in Commercial Hevea Natural Rubber by 2D-NMR with DOSY Filters and Multiple-WET Methods Using Ultrahigh-Field NMR.
Biomacromolecules
20(3), 1394-1400 (2019)
doi: 10.1021/acs.biomac.8b01771
Shi X, Prasanna C, Nagashima T, et al.
Structure and Dynamics in the Nucleosome Revealed by Solid-State NMR.
Angewandte Chemie
57(31), 9734-9738 (2018)
doi: 10.1002/anie.201804707
Yoo BK, Xiao Y, McElheny D, Ishii Y.
E22G Pathogenic Mutation of β-Amyloid (Aβ) Enhances Misfolding of Aβ40 by Unexpected Prion-like Cross Talk between Aβ42 and Aβ40.
Journal of the American Chemical Society
140(8), 2781-2784 (2018)
doi: 10.1021/jacs.7b13660
Xiao Y, Ma B, McElheny D, et al.
Aβ(1-42) fibril structure illuminates self-recognition and replication of amyloid in Alzheimer's disease.
Nature Structural & Molecular Biology
22(6), 499-505 (2015)
doi: 10.1038/nsmb.2991
Parthasarathy S, Inoue M, Xiao Y, et al.
Structural Insight into an Alzheimer's Brain-Derived Spherical Assembly of Amyloid β by Solid-State NMR.
Journal of the American Chemical Society
137(20), 6480-3 (2015)
doi: 10.1021/jacs.5b03373
Wickramasinghe NP, Parthasarathy S, Jones CR, et al.
Nanomole-scale protein solid-state NMR by breaking intrinsic 1HT1 boundaries.
Nature Methods
6(3), 215-8 (2009)
doi: 10.1038/nmeth.1300
メンバー
小林 直宏
上級研究員
髙橋 雅人
上級研究員
大山 貴子
研究員
WIJALATH PELIGE Ayesha
研究員
清宮 恭子
テクニカルスタッフⅠ
ニュース
2024年8月30日 研究成果
家族性アルツハイマー病に生じる特異な老人斑の謎に迫る
2024年4月15日 研究成果
SARS-CoV-2の転写開始の鍵となるRNA構造を同定

2024年1月5日 研究成果
複雑な形状を持つタンパク質をゼロから人工設計することに成功
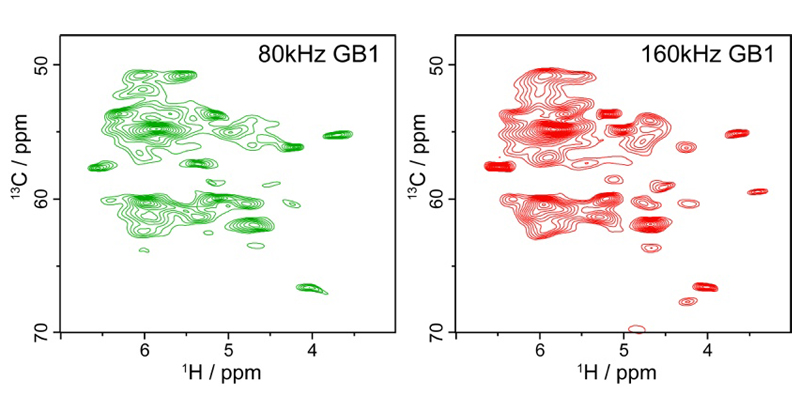
2023年7月24日 研究成果
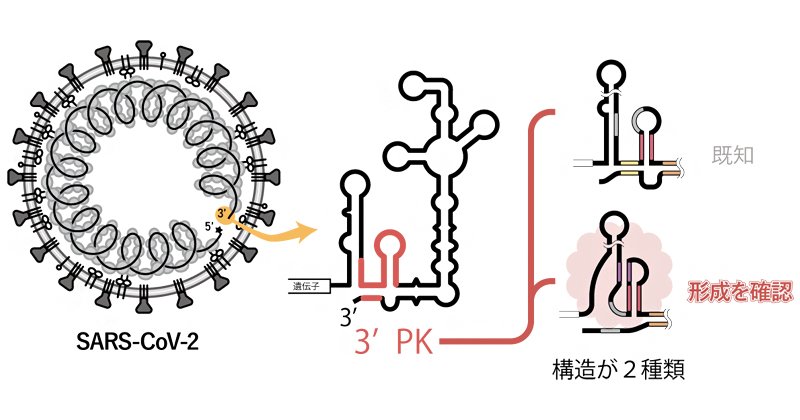
世界最高速で試料回転を行う固体NMRプローブを開発

2023年7月12日 研究成果
自然が見出していない未踏のタンパク質トポロジー

2022年11月25日 研究成果
卵子形成に必須なタンパク質DPPA3によるUHRF1の機能阻害の分子機構を解明
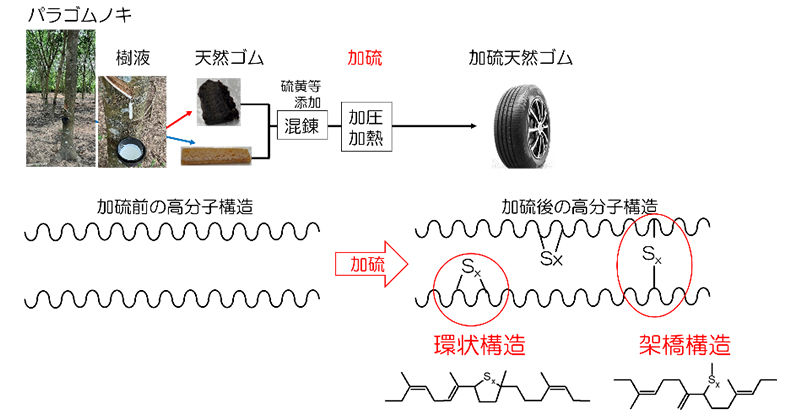
2022年10月26日 研究成果
加硫天然ゴム中の未知構造が明らかに
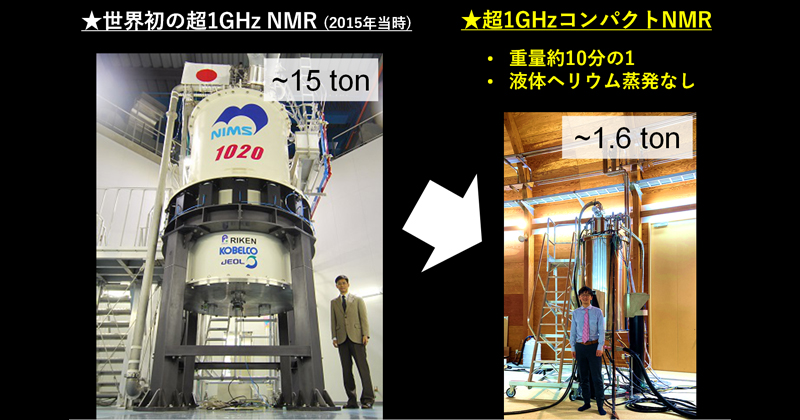
2022年10月25日 研究成果
世界一コンパクトな超1GHzのNMR装置の開発に成功

2021年9月24日 研究成果
電気抵抗のない高温超電導接合で2年間の永久電流運転に世界で初めて成功

2021年7月28日 研究成果
アルツハイマー病のタンパク質凝集体の構造の違いに迫る

2021年4月12日 BDRニュース
3チーム2ユニットが新たにBDRに加わりました!

2020年11月24日 研究成果
疎水性パッキングがゆるくても折り畳み能を示し超安定な人工タンパク質
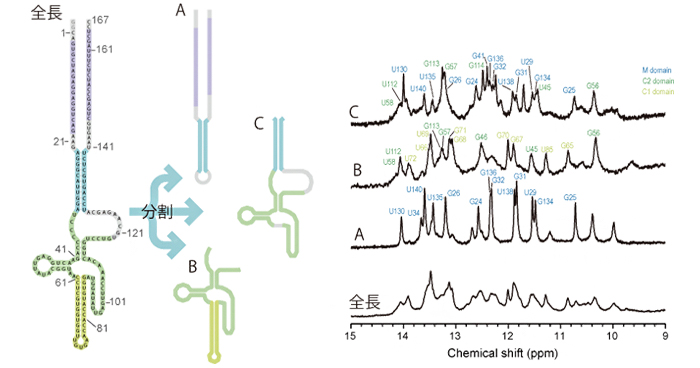
2020年8月6日 研究成果
タンパク質を増やす秘訣に迫る
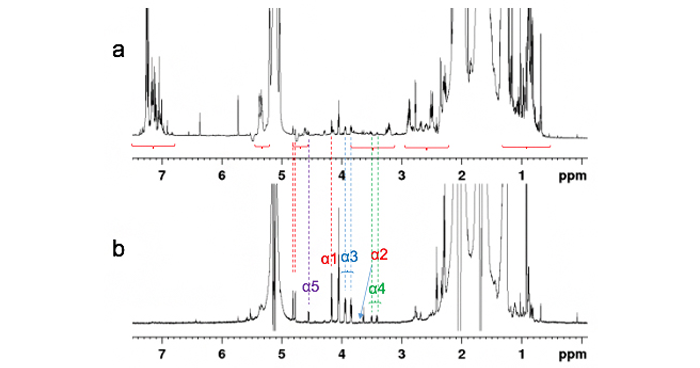
2019年3月11日 研究成果
新NMR測定法による天然ゴム末端基の解析