Creating an artificial genetic code
Mar. 18, 2021
The simplest cipher is created by using an encryption table (code) to convert one character into another. If we know the code, then the original message can be easily deciphered. Therefore, it is common to change the code frequently to make it more difficult to decipher. Over 50 years ago, biologists discovered the code for deciphering the genetic information of living organisms, and nowadays, they are capable of developing technologies to change the code itself.
The biological equivalent of a code is transfer RNA (tRNA), which plays a key role in "translating" the genetic information from messenger RNA (mRNA) into the amino acid sequences making up a protein. Team leader Yoshihiro Shimizu of RIKEN BDR’s Laboratory for Cell-Free Protein Synthesis, in collaboration with Takuya Ueda of The University of Tokyo and other colleagues, has succeeded in reconstructing an in vitro translation machinery using only artificially generated tRNAs. Furthermore, they showed that it is possible to create a genetic code that does not exist in nature by modifying the tRNAs. Their results were published in the journal, Communications Biology.
The sequences of the four mRNA nucleobases —adenine(A), cytosine (C), guanine (G), and uracil (U) —are grouped into sequences of three bases called codons. The mRNA codon is recognized by an anticodon, a three-nucleobase sequence, of the tRNA, which also carries the amino acid corresponding to this codon. The elongating end of the peptide strand is transferred to the amino acid carried by the tRNA, resulting in the elongation and folding of the peptide strand to generate a functional protein. In this way, the gene sequence is "translated" (or cryptographically decoded) into the amino acid sequence of the protein. Thus, we can say that tRNA is the entity of the code that converts the A-C-G-U mRNA sequences into amino acids (Figure 1A).
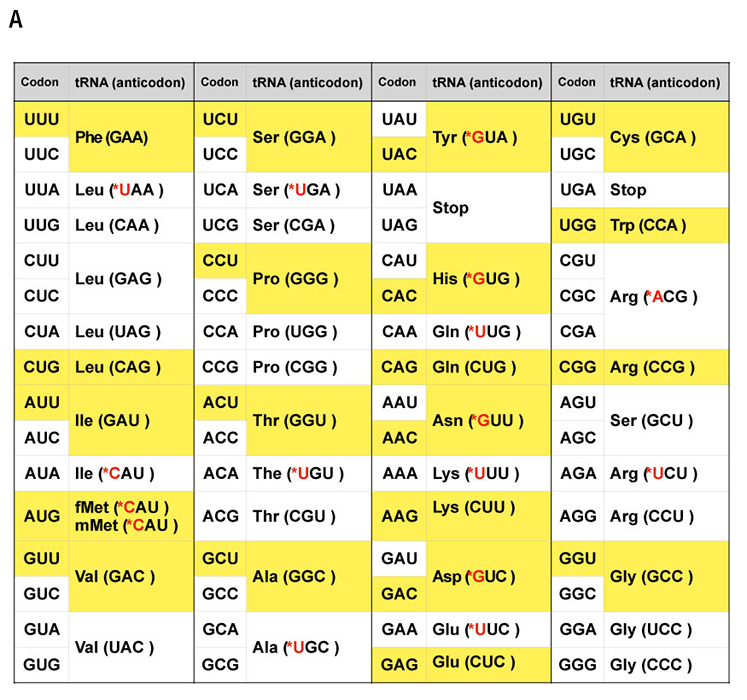
Some tRNAs recognize multiple codons through ambiguous multiple pairings between the third base in the codon and the first base in the anticodon, and in some cases, more than one tRNA species with different anticodons carry the same amino acid. The genetic code of E. coli consists of 41 tRNA species (Figure 1A).
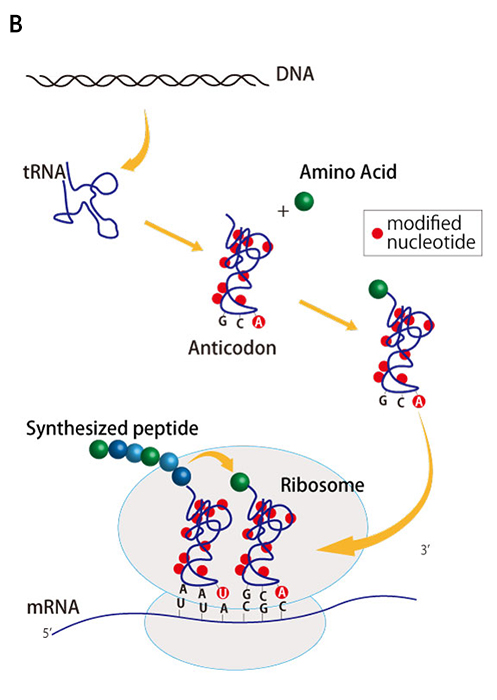
The research group previously developed an artificial translation system, named the PURE system, allowing them to synthesize proteins in vitro by purifying and mixing together all of the proteins and RNA complexes involved in translation in E. coli. However, this artificial translation system does not work when incorporating in vitro transcribed tRNAs instead of natural tRNAs. This is because the bases that make up natural tRNAs contain chemically modified bases, such as methylation, around the anticodon site, and in some cases, these modifications are essential for codon recognition (Figure 1B). Because some of these modifications are not easy to replicate in vitro, the research team hypothesized that if they selected those tRNAs that could either function or showed relatively little loss of function without the chemical modifications, they may be able to successfully reconstitute protein synthesis with their PURE system using only in vitro transcribed tRNAs.
They found that 21 in vitro transcribed tRNAs that functioned without modifications could be combined with the PURE system instead of natural tRNAs to translate all amino acid species (Figure 1A yellow cells). By preparing a DNA template for dihydrofolate reductase (DHFR), an enzyme that converts dihydrofolic acid to tetrahydrofolic acid, based on this minimal genetic code where each amino acid corresponds to only a single codon, the researchers were able to successfully demonstrate the generation of an active DHFR enzyme protein in vitro.
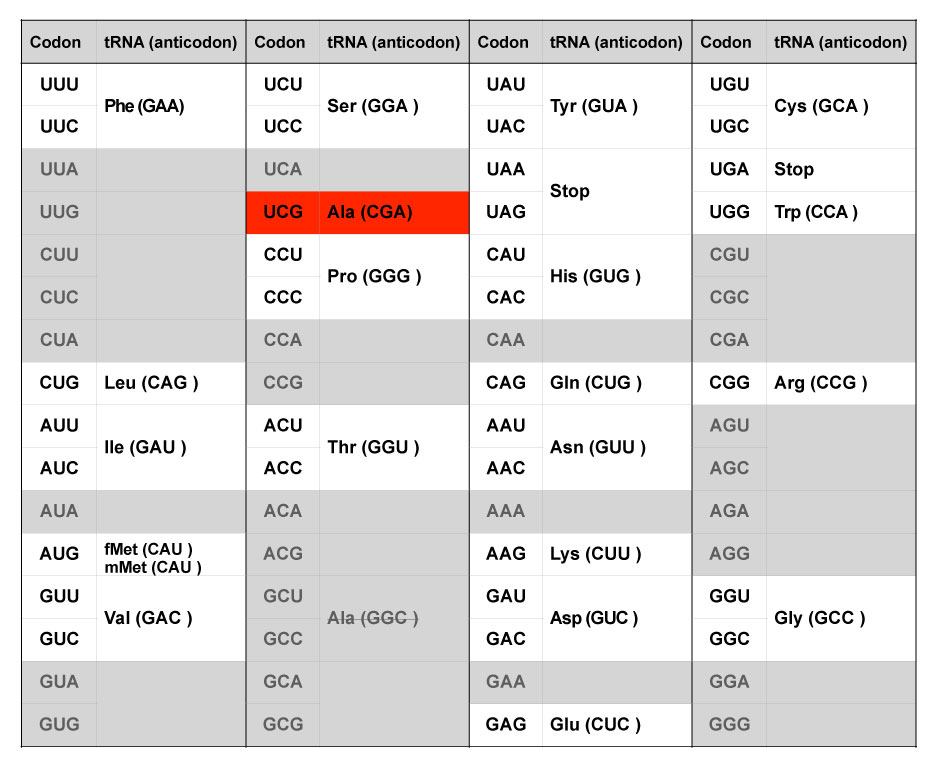
Codon UCG (corresponding to Ser in the native system) is assigned to Ala and tRNA containing the GGC anticodon (corresponding to Ala in the native system) is eliminated from the system.
The group also showed that they could experimentally overwrite the relationship between the tRNA anticodon sequence and amino acids. The anticodon of alanine tRNA, GGC, was replaced by the unused anticodon of Ser-tRNA, CGA. As a result, codon UCG (corresponding to anticodon CGA), which is recognized as serine (Ser) in the natural system, is interpreted as alanine (Ala) in this artificial system (Figure 2). They prepared another DNA template for DHFRs based on their newly designed genetic code, and when added to the PURE system with in vitro transcribed tRNAs, were able to successfully generate functional DHFRs. Using the same DNA template, they also showed that the system with a natural tRNA mixture produced an inactive DHFR protein because of the improper insertion of Ser instead of Ala.
With the COVID-19 pandemic, studies of coronaviruses and their proteins are urgently needed, but so are studies of dangerous viral proteins. The experimental system developed in this current study holds the potential for useful applications in studying them. When viral genes are modified by incorporating artificial genetic codes and their proteins are synthesized in vitro for research, even if the DNA is accidentally leaked into the environment and taken up by natural microbes and viruses, the proteins they would produce are likely to be inactive, and hence the risk to the environment is expected to be small.
"We think we can design an experimental system in which disease-causing genes, such as coronavirus proteins, can be completely detoxified outside the lab," says Shimizu on the future of the research. "I believe that using a cell-free system will prevent contamination of the biological environment, in the sense of not creating recombinant organisms."
By Takehiro Kawano
Publication Information
Hibi K, Amikura K, Sugiura N, et al.
Reconstituted cell-free protein synthesis using in vitro transcribed tRNAs.
Communications Biology 3, 350 (2020) doi: 10.1038/s42003-020-1074-2



