Team Leader
Kam Zhang
Ph.D.
Laboratory for Structural Bioinformatics
[Closed Mar. 2023]
E-mailkamzhang[at]riken.jp
Please replace [at] with @.
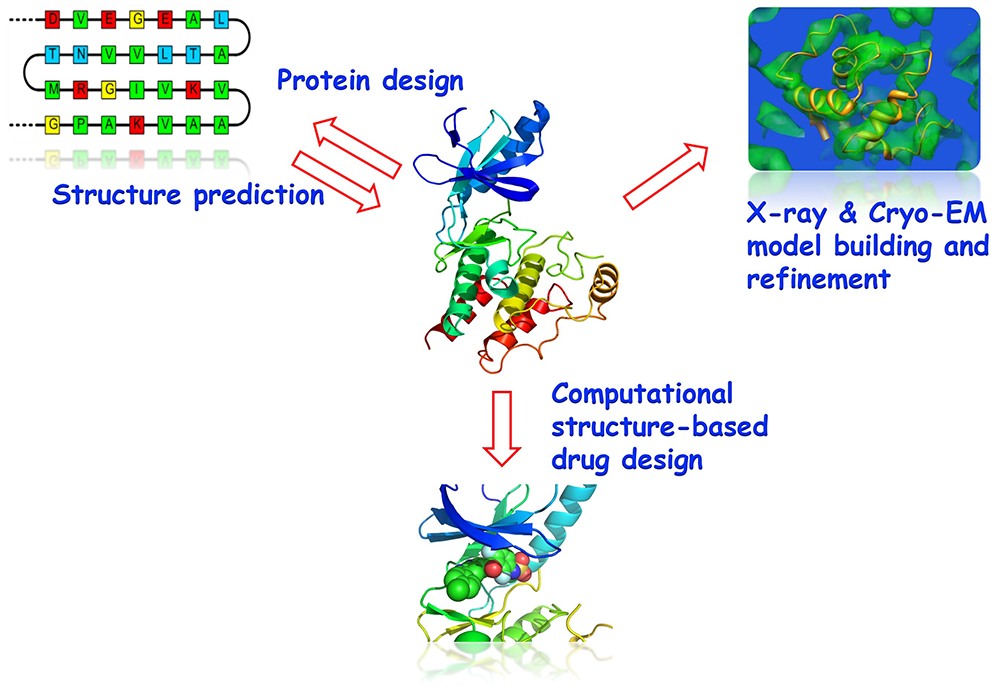
The complex biological functions of proteins are determined by their equally intricate three-dimensional structures. The correctly folded native structure is critical for the proper function of a protein in a cell. Small deviations from its native structure can often lead to malfunction of the protein and cause diseases. Our goal is to understand and modulate protein functions through computational studies of their structures. We are developing methods for protein structure prediction and applying design principles to create proteins with novel architectures, new biological functions or effective therapeutics. We are developing new methods of model building and refinement for X-ray and Cryo-EM. We are also developing and applying computational methods to discover new inhibitors for various drug targets.
Research Theme
- Protein structure prediction and design
- X-ray and Cryo-EM model building and refinement
- Virtual screening and drug design
Selected Publications
Alfi A, Popov A, Kumar A, et al.
Cell-Free Mutant Analysis Combined with Structure Prediction of a Lasso Peptide Biosynthetic Protease B2.
ACS Synthetic Biology
11(6), 2022-2028 (2022)
doi: 10.1021/acssynbio.2c00176
Yagi S, Padhi AK, Vucinic J, et al.
Seven Amino Acid Types Suffice to Create the Core Fold of RNA Polymerase.
Journal of the American Chemical Society
(2021)
doi: 10.1021/jacs.1c05367
Kaushik R, Zhang KYJ.
ProFitFun: A Protein Tertiary Structure Fitness Function for Quantifying the Accuracies of Model Structures.
Bioinformatics (Oxford, England)
(2021)
doi: 10.1093/bioinformatics/btab666
Padhi AK, Kumar A, Haruna KI, et al.
An integrated computational pipeline for designing high-affinity nanobodies with expanded genetic codes.
Briefings in bioinformatics
(2021)
doi: 10.1093/bib/bbab338
Jiang X, Kumar A, Motomura Y, et al.
A Series of Compounds Bearing a Dipyrido-Pyrimidine Scaffold Acting as Novel Human and Insect Pest Chitinase Inhibitors.
Journal of medicinal chemistry
63(3), 987-1001 (2020)
doi: 10.1021/acs.jmedchem.9b01154
Terada D, Voet ARD, Noguchi H, et al.
Computational design of a symmetrical ß-trefoil lectin with cancer cell binding activity.
Scientific Reports
7, 5943 (2017)
doi: 10.1038/s41598-017-06332-7
Voet ARD, Noguchi H, Addy C, et al.
Biomineralization of a Cadmium Chloride Nanocrystal by a Designed Symmetrical Protein.
Angewandte Chemie International Edition
54, 9857-9860 (2015)
doi: 10.1002/anie.201503575
Voet ARD, Noguchi H, Addy C, et al.
Computational design of a self-assembling symmetrical β-propeller protein.
Proceedings of the National Academy of Sciences of United States of America
111, 15102-15107 (2014)
doi: 10.1073/pnas.1412768111
Kumar A, Ito A, Hirohama M, et al.
Identification of Sumoylation Inhibitors Targeting a Predicted Pocket in Ubc9.
Journal of Chemical Information and Modeling
54, 2784−2793 (2014)
doi: 10.1021/ci5004015
Simoncini D, Zhang KYJ.
Efficient sampling in fragment-based protein structure prediction using an estimation of distribution algorithm.
PLoS ONE
8, e68954 (2013)
doi: 10.1371/journal.pone.0068954
Berenger F, Zhou Y, Shrestha R, Zhang KYJ.
Entropy-accelerated exact clustering of protein decoys.
Bioinformatics
27, 939 (2011)
doi: 10.1093/bioinformatics/btr072
Members

Team LeaderKam Zhang
- kamzhang[at]riken.jp
(Please replace [at] with @)

Senior ScientistAshutosh Kumar
- akumar[at]riken.jp

Research ScientistRahul Kaushik
- rahul.kaushik[at]riken.jp

Visiting ResearcherAditya Padhi
- adityakumar.padhi[at]riken.jp

International Program AssociateChun Lai Tam
- chunlai.tam[at]riken.jp
Student TraineeErma Fatiha Binti Muhammad
Visiting ScientistDileep Kalarickal Vijayan
Visiting ScientistFrancois Berenger
(Please replace [at] with @)