Team Leader
Hiroki Ueda
M.D., Ph.D.
Laboratory for Synthetic Biology
[Closed Mar. 2025]
E-mailuedah-tky@umin.ac.jp
A paradigm shift in biological research from reductionistic to synthetic approach has been undertaken in recent years. Even though the synthetic approach has been acknowledged as a central strategy of life science over the next few decades, basic technologies required in the synthetic approach, such like designing, controlling and manipulating biological molecules (DNAs, proteins and lipids) is remained to be established. The main tasks for laboratory for synthetic biology are to develop basic technologies for synthetic biology through close collaborative study within this center and to present pioneer studies in this new field.
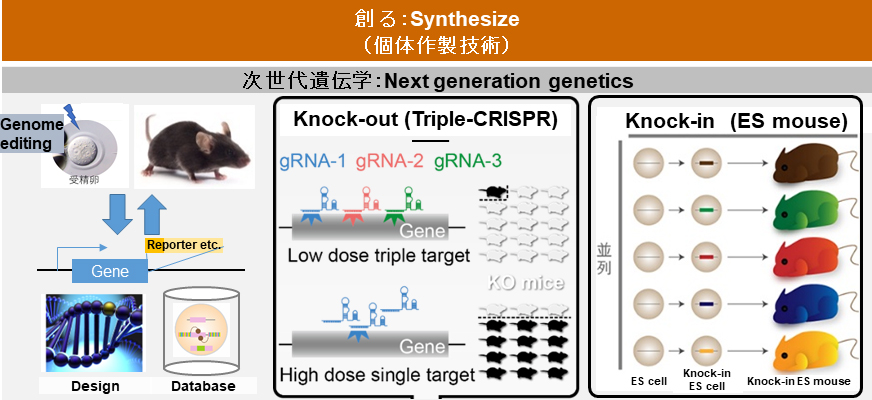
In our laboratory, we are developing innovative techniques to answer biological questions through the interdisciplinary efforts of collaborating members from multiple backgrounds. We have achieved several breakthroughs. (Top: Synthesize) Toward understanding biological systems at the organism level, we facilitate genetics by using information technology and developing efficient methods to produce genetically modified mice. Next generation genetics does not require the crossing that conventional methods do and allows researchers to prepare multiple lines of genetically modified mice in parallel within just a couple of months (left). The triple-target CRISPR method can produce whole-body biallelic knockout (KO) mice at high-efficiently (>90%) in a single generation (middle). The ES mouse method is a highly efficient procedure for producing knock-in (KI) mice (right).

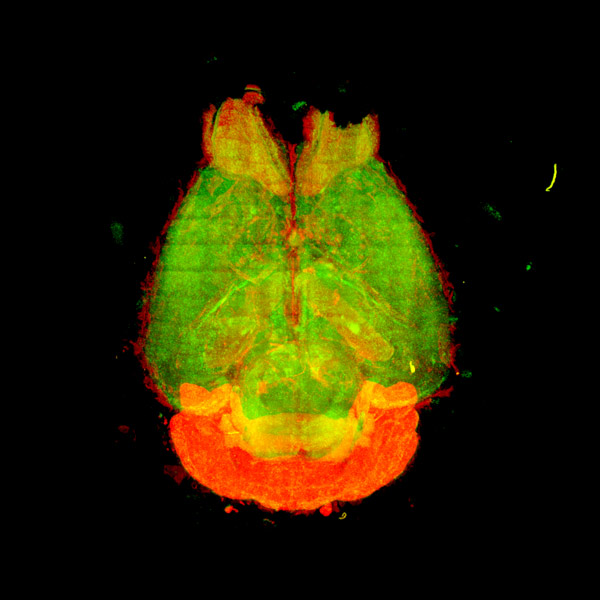
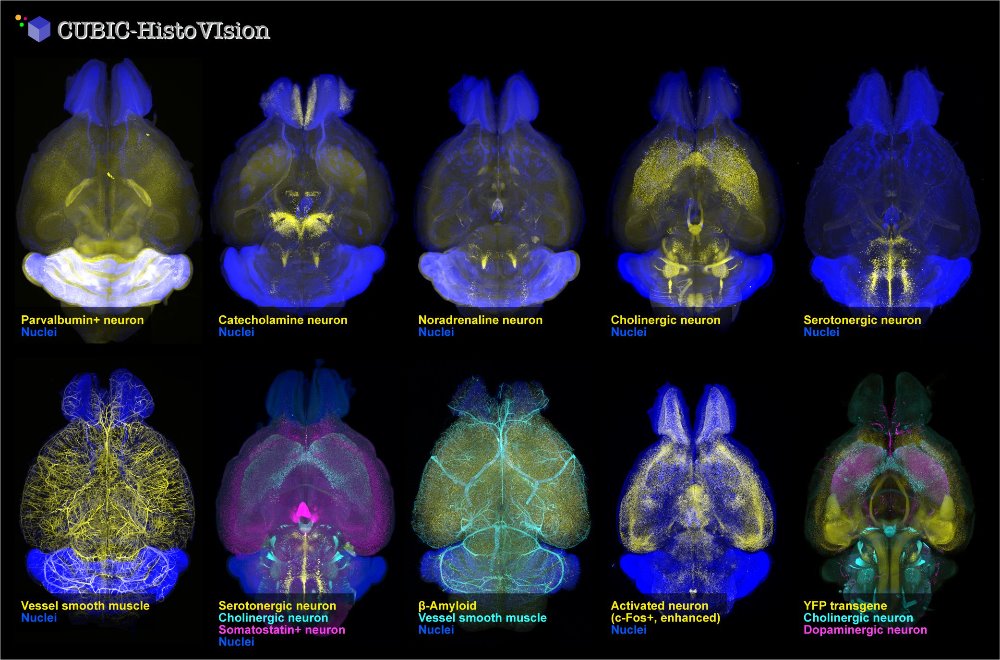
(left: Measure) The non-invasive respiration-based automated sleep phenotyping system – snappy sleep stager (SSS) – significantly simplifies and enhances the sleep phenotyping for which conventional methods required intricate and laborious work of a skilled researcher. (middle: See through) The tissue-clearing technique: CUBIC method innovates the way to observe deep inside of intact tissues by making them transparent. Example tissues shown above are brain, heart, liver, and kidney of mouse (from left to right). (right: Observe) The observation of transparent tissues is facilitated by a new technology of microscopy: light-sheet fluorescence microscopy (LSFM).
Research Theme
- Design of chemicals that can control and create dynamic protein behaviour.
- Design of molecular interactions in circadian clocks.
- Development of high-throughput genome engineering system.
- Gene synthesis for the design and construction of biological systems.
- Design and measurement of protein network dynamics at a single-cell level
Selected Publications
Susaki EA, Shimizu C, Kuno A, et al.
Versatile whole-organ/body staining and imaging based on electrolyte-gel properties of biological tissues.
Nature Communications
11, 1982 (2020)
doi: 10.1038/s41467-020-15906-5
Funano SI, Tone D, Ukai H, et al.
Rapid and easy-to-use ES cell manipulation device with a small groove near culturing wells.
BMC research notes
13(1), 453 (2020)
doi: 10.1186/s13104-020-05294-w
Ota N, Kanda GN, Moriguchi H, et al.
A Microfluidic Platform Based on Robust Gas and Liquid Exchange for Long-term Culturing of Explanted Tissues.
Analytical sciences : the international journal of the Japan Society for Analytical Chemistry
35(10), 1141-1147 (2019)
doi: 10.2116/analsci.19P099
Matsumoto K, Mitani TT, Horiguchi SA, et al.
Advanced CUBIC tissue clearing for whole-organ cell profiling.
Nature Protocols
14, 3506-3537 (2019)
doi: 10.1038/s41596-019-0240-9
Niwa Y, Kanda GN, Yamada RG, et al.
Muscarinic Acetylcholine Receptors Chrm1 and Chrm3 Are Essential for REM Sleep.
Cell reports
24(9), 2231-2247 (2018)
doi: 10.1016/j.celrep.2018.07.082
Tainaka K, Murakami TC, Susaki EA, et al.
Chemical Landscape for Tissue Clearing Based on Hydrophilic Reagents.
Cell reports
24(8), 2196-2210 (2018)
doi: 10.1016/j.celrep.2018.07.056
Ukai H, Kiyonari H, Ueda HR.
Production of knock-in mice in a single generation from embryonic stem cells.
Nature Protocol
12, 2513-2530 (2017)
doi: 10.1038/nprot.2017.110
Shinohara Y, Koyama YM, Ukai-Tadenuma M et al.
Temperature-Sensitive Substrate and Product Binding Underlie Temperature-Compensated Phosphorylation in the Clock.
Molecular Cell
67, 783-798 (2017)
doi: 10.1016/j.molcel.2017.08.009
Kubota SI, Takahashi K, et al.
Whole-Body Profiling of Cancer Metastasis with Single-Cell Resolution.
Cell Reports
20, 236-250 (2017)
doi: 10.1016/j.celrep.2017.06.010
Ode KL, Ukai H, Susaki EA, et al.
Knockout-rescue embryonic stem cell-derived mouse reveals circadian-period control by quality and quantity of CRY1.
Molecular Cell
65, 176-190 (2017)
doi: 10.1016/j.molcel.2016.11.022
Narumi R, Shimizu Y, et al.
Mass spectrometry-based absolute quantification reveals rhythmic variation of mouse circadian clock proteins.
Proceedings National Academy of Sciences of the United States of America
133, E3461-E3467 (2016)
doi: 10.1073/pnas.1603799113
Tatsuki F, Sunagawa GA, Shi S, et al.
Involvement of Ca2+-Dependent Hyperpolarization in Sleep Duration in Mammals.
Neuron
90(1), 70-85 (2016)
doi: 10.1016/j.neuron.2016.02.032
Sunagawa GA.
Mammalian Reverse Genetics without Crossing Reveals Nr3a as a Short-Sleeper Gene.
Cell Reports
14(3), 662-677 (2016)
doi: 10.1016/j.celrep.2015.12.052
Susaki EA, Tainaka K, Perrin D, et al.
Advanced CUBIC protocols for whole-brain and whole-body clearing and imaging.
Nature Protocol
10(11), 1709-1727 (2015)
doi: 10.1038/nprot.2015.085
Tainaka K, Kubota SI, Suyama TQ, et al.
Whole-Body Imaging with Single-Cell Resolution by Tissue Decolorization.
Cell
159(4), 911-924 (2014)
doi: 10.1016/j.cell.2014.10.034
Susaki EA, Tainaka K, Perrin D, et al.
Whole-Brain Imaging with Single-Cell Resolution Using Chemical Cocktails and Computational Analysis.
Cell
157(3), 726-739 (2014)
doi: 10.1016/j.cell.2014.03.042
Members
Rikuhiro Yamada
Senior Research Scientist
Hideki Ukai
Senior Research Scientist
Yohei Koyama
Research Scientist
Katsuhiko Matsumoto
Research Scientist
Genki Kanda
Special Postdoctoral Researcher
Hiroko Yukinaga
Special Postdoctoral Researcher
Arthur Robert Millius
Foreign Postdoctoral Researcher
Yuta Shinohara
Postdoctoral Researcher
Daisuke Tone
Postdoctoral Researcher
Maki Ukai
Technical Staff I
Hiroshi Fujishima
Technical Staff I
Chika Shimizu
Technical Staff I
Junko Garcon
Technical Staff I