Team Leader
Yuichi Taniguchi
Ph.D.
Laboratory for Cell Systems Control
[Closed Mar. 2023]
E-mailtaniguchi[at]riken.jp
Please replace [at] with @.
We are working to reveal rules and logic of single cell systems dynamics by developing innovative technologies. We aim to explore comprehensive and exhaustive properties of single cells based on cutting-edge technologies including single molecule imaging, high-throughput analysis, 3D genome structure analysis, molecular dynamics simulation, machine learning, MEMS and bioinformatics. We also intend to create new concepts for describing and understanding complex behaviors of single cells regulated by vast numbers of genes and bio-molecules. Ultimately, we aim to provide key principles for forecasting and controlling complex cell behaviors.
Research Theme
- Establishment of single-molecule imaging methods for detecting biomolecules in single cells.
- Development of methods for comprehensive and exhaustive characterization of the cell.
- Exploring regulatory mechanisms of the cell based on genome structure and epigenome analyses.
- Modeling rules and logic for complex single cell systems dynamics.
- Development of methods for controlling cellular states and medical applications.
Selected Publications
Ohno M, Ando T, Priest DG, Taniguchi Y.
Hi-CO: 3D genome structure analysis with nucleosome resolution.
Nature Protocols
16(7), 3439-3469 (2021)
doi: 10.1038/s41596-021-00543-z
Shinkai S, Nakagawa M, Sugawara T, et al.
PHi-C: deciphering Hi-C data into polymer dynamics.
NAR Genomics and Bioinformatics
2, lqaa020 (2020)
doi: 10.1093/nargab/lqaa020
Kumar V, Leclerc S, Taniguchi Y.
BHi-Cect: A top-down algorithm for identifying the multi-scale hierarchical structure of chromosomes.
Nucleic Acids Research
48, e26 (2020)
doi: https://academic.oup.com/nar/advance-article/doi/10.1093/nar/gkaa004/5713458
Yoshida Y, Taniguchi Y.
Simultaneous Single-Cell Measurements Demonstrate a Positive Correlation between RNA Copy Number for Mitochondrial Division and Fusion Genes and Mitochondrial Fragmentation.
Cytologia
84(1), 15-23 (2019)
doi: 10.1508/cytologia.84.15
Ohno M, Ando T, Priest D.G, et al.
Sub-nucleosomal genome structure reveals distinct nucleosome folding motifs.
Cell
176(3), 520-534.e25 (2019)
doi: 10.1016/j.cell.2018.12.014
Leclerc S, Arntz Y, Taniguchi Y.
Proteome-wide Quantification of Labeling Homogeneity at the Single Molecule Level.
Journal of visualized experiments : JoVE
(2019)
doi: 10.3791/59199
Leclerc S, Arntz Y, Taniguchi Y.
Extending Single Molecule Imaging to Proteome Analysis by Quantitation of Fluorescent Labeling Homogeneity in Complex Protein Samples.
Bioconjugate chemistry
29(8), 2541-2549 (2018)
doi: 10.1021/acs.bioconjchem.8b00226
Ohno M, Priest DG, Taniguchi Y.
Nucleosome-level 3D organization of the genome.
Biochemical Society transactions
46(3), 491-501 (2018)
doi: 10.1042/BST20170388
Priest D, Tanaka N, Tanaka Y, Taniguchi Y.
Micro-patterned agarose gel devices for single-cell high-throughput microscopy of E. coli cells.
Scientific Reports
7, 17750 (2017)
doi: 10.1038/s41598-017-17544-2
Taniguchi Y.
Autofluorescence imaging of tissue samples using super-high sensitivity fluorescence microscopy.
Global Imaging Insights
2, 1-2 (2017)
doi: 10.15761/GII.1000135
Taniguchi Y.
Genome-Wide Analysis of Protein and mRNA Copy Numbers in Single Escherichia coli Cells with Single-Molecule Sensitivity.
Methods in Molecular Biology
1346, 55-67 (2015)
doi: 10.1007/978-1-4939-2987-0_5
Ohno M, Karagiannis P, Taniguchi Y.
Protein Expression Analyses at the Single Cell Level.
Molecules
19(9), 13932-13947 (2014)
doi: 10.3390/molecules190913932
Taniguchi Y, Choi PJ, Li GW, et al.
Quantifying E-coli Proteome and Transcriptome with Single-Molecule Sensitivity in Single Cells.
Science
329(5991), 533-538 (2010)
doi: 10.1126/science.1188308
Taniguchi Y, Nishiyama M, Ishii Y, Yanagida T.
Entropy rectifies the Brownian steps of kinesin.
Nature Chem. Biol.
1, 342-347 (2005)
Members
Masae Ohno Johmura
Visiting Scientist
Soo Yeon Kim
Special Postdoctoral Researcher
Mayumi Ishida
Technical Staff I
Kaori Tsujimura
Technical Staff I
Binti Kamarulzaman Latiefa
Student Trainee
Shouko Ohara
Administrative Part-time Worker II
Yuriko Okuda
Assistant
News

Apr. 28, 2022 BDR News
Four BDR scientists awarded MEXT prizes

Sep. 30, 2021 BDR News
BDR team leader awarded the Osaka Science Prize

Jun. 2, 2020 BDR News
Dive into BDR's intriguing research
Computational work to see the structure of DNA
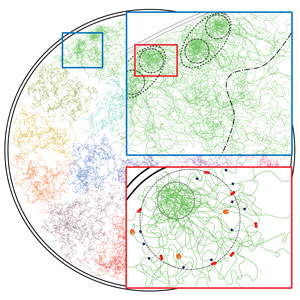
May 1, 2020 Research
Finding more chromosome structures by assuming less
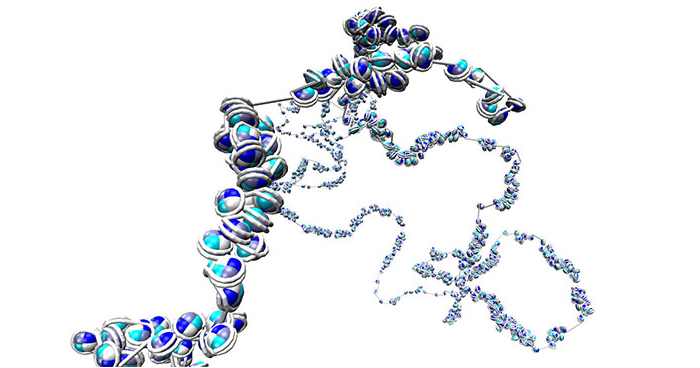
Jul. 5, 2019 Research
Analyzing chromatin structure in more detail than ever before



