
Team Leader
Kenta Sumiyama*
Ph.D.
Laboratory for Mouse Genetic Engineering
[Closed Sep. 2023]
*Current position
Professor, Graduate School of Bioagricultural Sciences, Nagoya University
emailksumiyam[at]agr.nagoya-u.ac.jp
Please replace [at] with @.
We are developing new techniques in mouse genetic engineering including direct genome editing on fertilized eggs of mice and microdevice-based easy and precise aggregation methods to obtain complete ES cell derived chimeric mice. By using these tools such as Triple CRISPR or Tol2 transgenesis, we are producing various useful genetically engineered mice within a short period of time. We are also studying cis-regulatory mechanisms in non-coding regions by using both molecular evolutionary analysis and genome editing technology to elucidate developmental and evolutionary mechanisms of mammals. Our recent achievement is the triple CRISPR gene knockout method, enabling 100% knockout of first generation mice. By using this method, phenotypic analysis can be done within three months without further crossing and breeding.
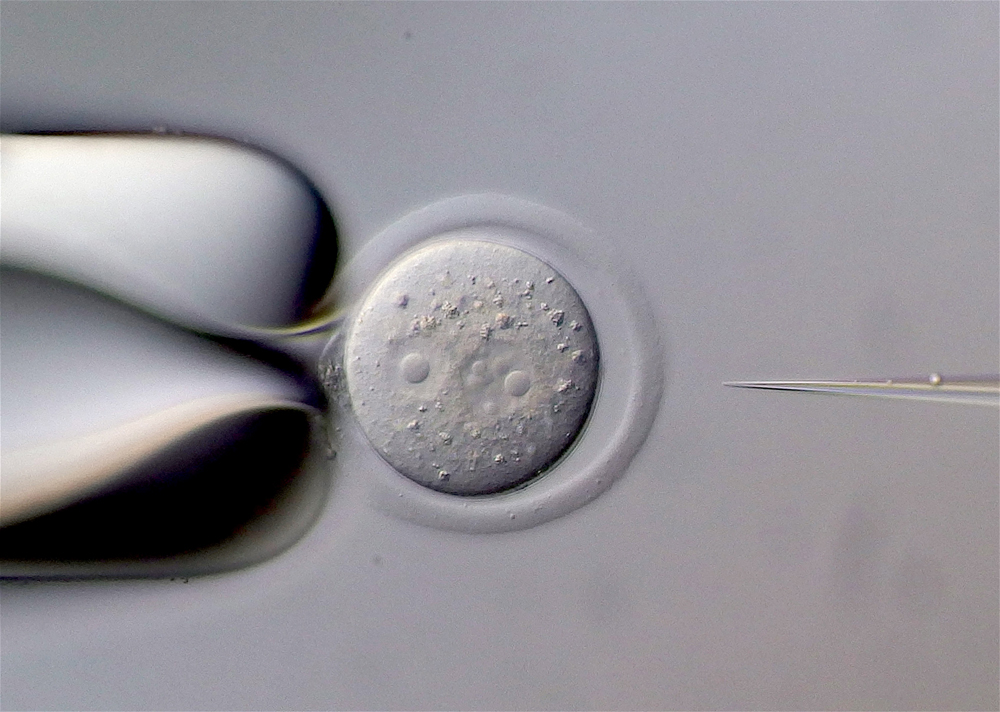
Microinjection into a mouse fertilized egg
Research Theme
- Developing new techniques in highly efficient direct genome editing on fertilized eggs of mice
- Developing new transgenesis techniques on fertilized eggs of mice
- Study of cis-regulatory function in non-coding region of the mammalian genome
- Study of evolutionary mechanisms for acquiring novel function
Selected Publications
Sumiyama K, Matsumoto N, Garçon-Yoshida J, et al.
Easy and efficient production of completely embryonic-stem-cell-derived mice using a micro-aggregation device.
PloS one
13(9), e0203056 (2018)
doi: 10.1371/journal.pone.0203056
Tatsuki F, Sunagawa GA, Shi S, et al.
Involvement of Ca2+-Dependent Hyperpolarization in Sleep Duration in Mammals.
Neuron
90(1), 70-85 (2016)
doi: 10.1016/j.neuron.2016.02.032
Sunagawa GA, Sumiyama K, Ukai-Tadenuma M, et al.
Mammalian Reverse Genetics without Crossing Reveals Nr3a as a Short-Sleeper Gene.
Cell Reports
14(3), 662-677 (2016)
doi: 10.1016/j.celrep.2015.12.052
Noguchi M, Sumiyama K, Morimoto M.
Directed Migration of Pulmonary Neuroendocrine Cells toward Airway Branches Organizes the Stereotypic Location of Neuroepithelial Bodies.
Cell Reports
13(12), 2679-2686 (2015)
doi: 10.1016/j.celrep.2015.11.058
Goto A, Nakahara I, Yamaguchi T, et al.
Circuit-dependent striatal PKA and ERK signaling underlies rapid behavioral shift in mating reaction of male mice.
Proceedings Of The National Academy Of Sciences Of The United States Of America
112(21), 6718-6723 (2015)
doi: 10.1073/pnas.1507121112
Amemiya CT, Alfoldi J, Lee AP, et al.
The African coelacanth genome provides insights into tetrapod evolution.
Nature
496(7445), 311-316 (2013)
doi: 10.1038/nature12027
Sumiyama K, Miyake T, Grimwood J, et al.
Theria-Specific Homeodomain and cis-Regulatory Element Evolution of the Dlx3-4 Bigene Cluster in 12 Different Mammalian Species.
Journal of Experimental Zoology Part B-Molecular and Developmental Evolution
318(8), 639-650 (2012)
doi: 10.1002/jez.b.22469
Kamioka Y, Sumiyama K, Mizuno R, et al.
Live Imaging of Protein Kinase Activities in Transgenic Mice Expressing FRET Biosensors.
Cell Structure and Function
37(1), 65-73 (2012)
Sumiyama K, Kawakami K, Yagita K.
A simple and highly efficient transgenesis method in mice with the Tol2 transposon system and cytoplasmic microinjection.
Genomics
95(5), 306-311 (2010)
doi: 10.1016/j.ygeno.2010.02.006
Sumiyama K, Ruddle FH.
Regulation of Dlx3 gene expression in visceral arches by evolutionarily conserved enhancer elements.
Proceedings of the National Academy of Sciences of the United States of America
100(7), 4030-4034 (2003)
doi: 10.1073/pnas.0530119100
Sumiyama K, Irvine SQ, Stock DW, et al.
Genomic structure and functional control of the Dlx3-7 bigene cluster.
Proceedings of the National Academy of Sciences of the United States of America
99(2), 780-785 (2002)
doi: 10.1073/pnas.012584999



