
Team Director
Mikako Shirouzu
Ph.D.
Laboratory for Protein Functional and Structural Biology
[Affiliation has changed to RIKEN Center for Integrated Medical Sciences (IMS) as of April 2025]
LocationYokohama
E-mailmikako.shirouzu@riken.jp
The high-resolution structural information of proteins related to diseases shall increasingly become important for drug development leading to future individualized medicine. We plan to establish a structural analysis technology platform to contribute to "Life innovation" such as drug discovery as well as development of methods for the sample preparation of challenging proteins including membrane proteins/biomolecular complexes and for the structural analysis by cryo-electron microscopy (cryo-EM). The 3D-structural information will be used for in-silico screening/design of chemical compounds and for dynamic structural analysis toward simulation research of cell function.
Research Theme
- Analysis of developmental process using theoretical cell models
- Study of evolutionary dynamics of microorganisms by comprehensive phenotypic/genetic analysis
- Development of algorithm for high-resolution comprehensive phenotypic/genetic analysis
Selected Publications
Kaneko S, Imai S, Uchikubo-Kamo T, et al.
Structural and dynamic insights into the activation of the μ-opioid receptor by an allosteric modulator.
Nature Communications
15(1), 3544 (2024)
doi: 10.1038/s41467-024-47792-6
Yamagata A, Ito K, Suzuki T, et al.
Structural basis for antiepileptic drugs and botulinum neurotoxin recognition of SV2A.
Nature Communications
15(1), 3027 (2024)
doi: 10.1038/s41467-024-47322-4
Akiyama N, Ishiguro K, Yokoyama T, et al.
Structural insights into the decoding capability of isoleucine tRNAs with lysidine and agmatidine.
Nature Structural & Molecular Biology
(2024)
doi: 10.1038/s41594-024-01238-1
Suzuki M, Uchibori K, Oh-Hara T, et al.
A macrocyclic kinase inhibitor overcomes triple resistant mutations in EGFR-positive lung cancer.
Npj Precision Oncology
8(1), 46 (2024)
doi: 10.1038/s41698-024-00542-9
Tanaka M, Yokoyama T, Saito H, et al.
Boric acid intercepts 80S ribosome migration from AUG-stop by stabilizing eRF1.
Nature Chemical Biology
(2024)
doi: 10.1038/s41589-023-01513-0
Furutani Y, Hirano Y, Toguchi M, et al.
A small molecule iCDM-34 identified by in silico screening suppresses HBV DNA through activation of aryl hydrocarbon receptor.
Cell Death Discovery
9(1), 467 (2023)
doi: 10.1038/s41420-023-01755-w
Zhao X, Ma D, Ishiguro K, et al.
Glycosylated queuosines in tRNAs optimize translational rate and post-embryonic growth.
Cell
186(25), 5517-5535.e24 (2023)
doi: 10.1016/j.cell.2023.10.026
Kikuchi M, Morita S, Wakamori M, et al.
Epigenetic mechanisms to propagate histone acetylation by p300/CBP.
Nature Communications
14(1), 4103 (2023)
doi: 10.1038/s41467-023-39735-4
Yamagata A, Murata Y, Namba K, et al.
Uptake mechanism of iron-phytosiderophore from the soil based on the structure of yellow stripe transporter.
Nature Communications
13, 7180 (2022)
doi: 10.1038/s41467-022-34930-1
Ehara H, Kujirai T, Shirouzu M, et al.
Structural basis of nucleosome disassembly and reassembly by RNAPII elongation complex with FACT.
Science
377(6611), eabp9466 (2022)
doi: 10.1126/science.abp9466
Nakamura H, Hisano T, Rahman MM, et al.
Structural basis for heme detoxification by an ATP-binding cassette-type efflux pump in gram-positive pathogenic bacteria.
Proceedings of the National Academy of Sciences of the United States of America
119(27), e2123385119 (2022)
doi: 10.1073/pnas.2123385119
Park JH, Iwamoto M, Yun JH, et al.
Structural insights into the HBV receptor and bile acid transporter NTCP.
Nature
606(7916), 1027-1031 (2022)
doi: 10.1038/s41586-022-04857-0
Hosaka T, Nomura T, Kubo M, et al.
Conformational alterations in unidirectional ion transport of a light-driven chloride pump revealed using X-ray free electron lasers.
Proceedings of the National Academy of Sciences of the United States of America
119(9), e2117433119 (2022)
doi: 10.1073/pnas.2117433119
Nakagawa Y, Shen HC, Komi Y, et al.
Amyloid conformation-dependent disaggregation in a reconstituted yeast prion system.
Nature Chemical Biology
18(3), 321-331 (2022)
doi: 10.1038/s41589-021-00951-y
Kasahara K, Re S, Nawrocki G, et al.
Reduced Efficacy of a Src Kinase Inhibitor in Crowded Protein Solution.
Nature Communications
12, 4099 (2021)
doi: 10.1038/s41467-021-24349-5
Kukimoto-Niino M, Katsura K, Kaushik R, et al.
Cryo-EM structure of the human ELMO1-DOCK5-Rac1 complex.
Science Advances
7(30), eabg3147 (2021)
doi: 10.1126/sciadv.abg3147
News
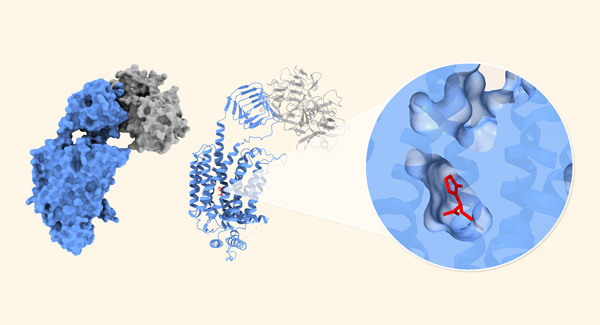
Sep. 27, 2024 Research
Structural secrets of antiepileptic drugs uncovered

Mar. 16, 2023 Research
A nifty trick to help plants thrive in iron-poor soils
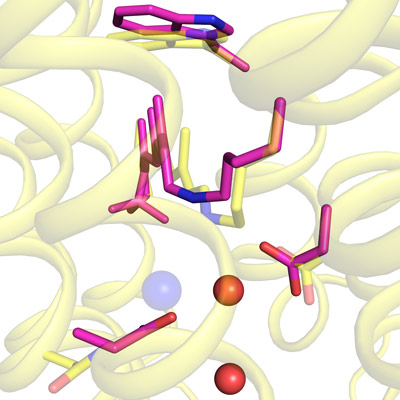
Jun. 9, 2022 Research
Imaging how a light-driven chloride pump works
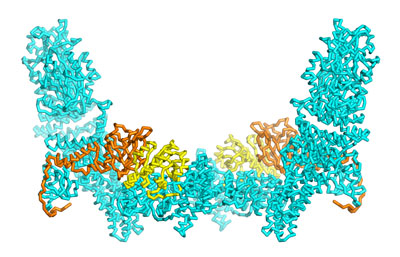
Nov. 1, 2021 Research
Mapping the structural features that regulate a signaling protein’s specificity
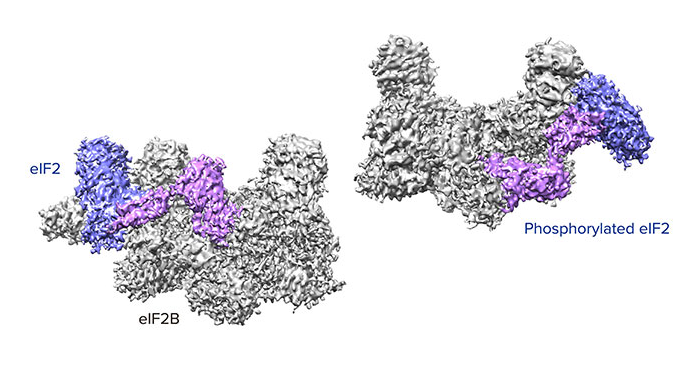
Aug. 9, 2019 Research
How phosphorylation of eIF2 reduces protein synthesis
Dec. 7, 2018 Research
Cryo-electron microscopy shows how stabilizing proteins bind to microtubules



