RIKEN ECL Team Leader
Masayuki Oginuma
Ph.D.
Chrono-Developmental Biology RIKEN ECL Research Team
LocationKobe / Developmental Biology Buildings
E-mailmasayuki.oginuma@riken.jp
Organisms possess a remarkable "blueprint of time." Embryonic development, from fertilization to the processes of body formation, follows a precise schedule regulated by a clock mechanism encoded in the genes. However, the specific underlying molecular mechanisms remains largely unknown. The turquoise killifish, N. furzeri, which inhabit the ephemeral ponds in Africa, can suspend their embryonic development and enter a state of diapause for extended periods.
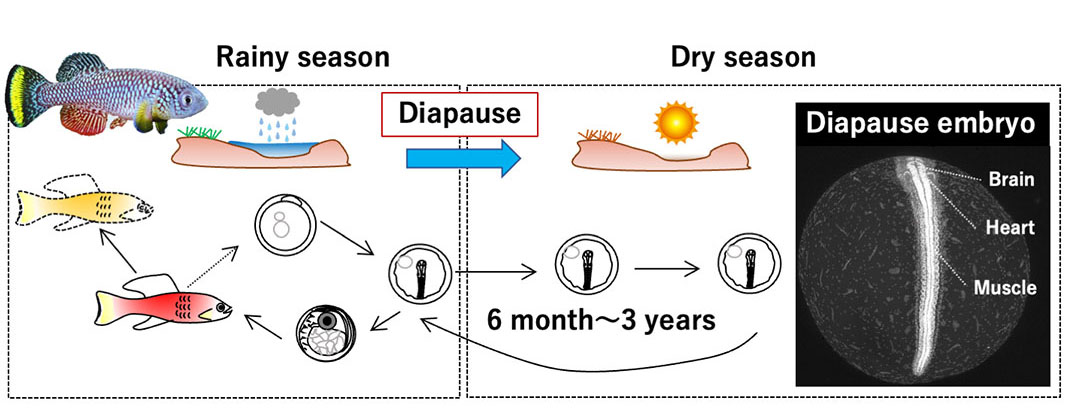 Figure 1. Life cycle of turquoise killifish
Figure 1. Life cycle of turquoise killifish
Our lab aims to clarify the mechanisms of diapause, a highly evolved developmental clock-stopping phenomenon. We have developed a method that allows us to rapidly and easily analyze the dynamics and functions of turquoise killifish genes, and using this method, we will investigate the function of genes that regulate embryonic time to reveal the complete picture of the "blueprint of time," and applying our findings to other organisms.
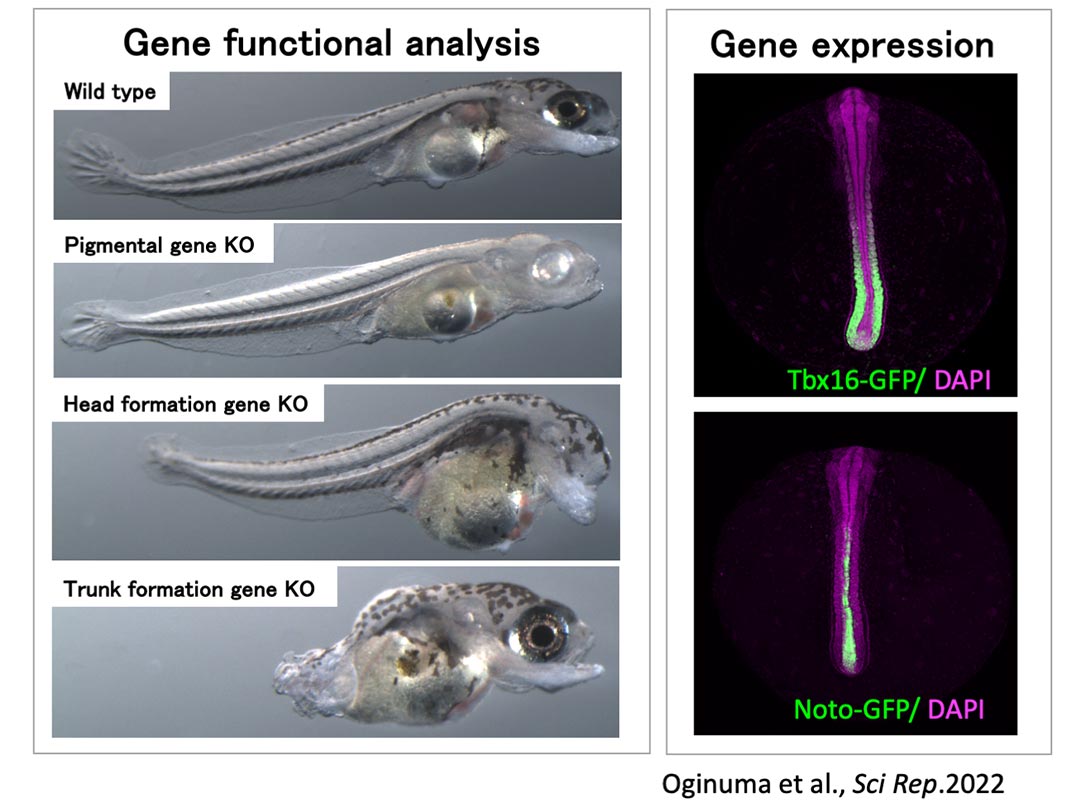 Figure 2. Turquoise killifish genetic analysis platform
Figure 2. Turquoise killifish genetic analysis platform
Selected Publications
Oginuma M, Nishida M, Ohmura-Adachi T, et al.
Rapid reverse genetics systems for Nothobranchius furzeri, a suitable model organism to study vertebrate aging.
Scientific Reports
12(1), 11628 (2022)
doi: 10.1038/s41598-022-15972-3
Oginuma M, Harima Y, Tarazona OA, et al.
Intracellular pH controls WNT downstream of glycolysis in amniote embryos.
Nature
584(7819), 98-101 (2020)
doi: 10.1038/s41586-020-2428-0
Zhao W, Oginuma M, Ajima R, et al.
Ripply2 recruits proteasome complex for Tbx6 degradation to define segment border during murine somitogenesis.
eLife
7, e33068 (2018)
doi: 10.7554/eLife.33068
Oginuma M, Moncuquet P, Xiong F, et al.
A Gradient of Glycolytic Activity Coordinates FGF and Wnt Signaling during Elongation of the Body Axis in Amniote Embryos.
Developmental Cell
40(4), 342-353 (2017)
doi: 10.1016/j.devcel.2017.02.001
Chal J, Oginuma M, Al Tanoury Z, et al.
Differentiation of pluripotent stem cells to muscle fiber to model Duchenne muscular dystrophy.
Nature Biotechnology
33(9), 962-969 (2015)
doi: 10.1038/nbt.3297
Oginuma M, Takahashi Y, Kitajima S, et al.
The oscillation of Notch activation, but not its boundary, is required for somite border formation and rostral-caudal patterning within a somite.
Development
137(9), 1515-1522 (2010)
doi: 10.1242/dev.044545
Oginuma M, Hirata T, Saga Y.
Identification of presomitic mesoderm (PSM)-specific Mesp1 enhancer and generation of a PSM-specific Mesp1/Mesp2-null mouse using BAC-based rescue technology.
Mechanisms of Development
125(5-6), 432-440 (2008)
doi: 10.1016/j.mod.2008.01.010
Oginuma M, Niwa Y, Chapman DL, Saga Y.
Mesp2 and Tbx6 cooperatively create periodic patterns coupled with the clock machinery during mouse somitogenesis.
Development
135(15), 2555-2562 (2008)
doi: 10.1242/dev.019877
Members
Masayuki Oginuma
RIKEN ECL Team Leader
Daniel Semmy
Postdoctoral Researcher
Mari Kaneko
Technical Staff I
Yuko Sakaide
Technical Staff I
Bhavana Venkatesh Murthy
Technical Staff II
Miki Fujiwara
Assistant
Aine Minato
Student Trainee
Nozomi Ugajin
Visiting Scientist