Team Leader
Takashi Nishimura
Ph.D.
Laboratory for Growth Control Signaling
[Closed Jun. 2019]
*Current email address : t-nishimura[at]gunma-u.ac.jp
Please replace [at] with @.
The processes of animal development, including organ size and body size, are genetically predetermined, but these processes are also influenced by environmental factors such as nutrition and temperature. The close link between cell and tissue growth control and environmental cues ensures that developmental transitions occur at the appropriate time during animal development.
Cell proliferation and differentiation in each tissue and organ are kept under strict regulation, both spatially and temporally. Research has revealed the nature of spatial signals, such as growth factors and morphogens, but the way in which these signals direct cell and tissue growth over time remains poorly understood. In addition, growth and developmental timing are also governed by nutrient availability. Most species have a standard body size, but developing organisms are also capable of adapting their growth to fluctuating nutritional states through metabolic regulation. Therefore, linking the nutrient sensing system to an endocrine signaling network allows organisms to control the timing of cell proliferation and differentiation.
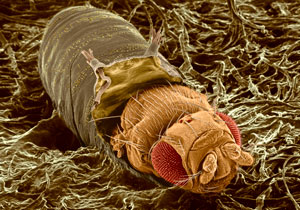
Our team's research aims to shed light on the molecular basis for growth control and developmental timing at the cellular and tissue/organ level using Drosophila as a model system. In particular, we are interested in addressing the following questions: 1) How do organisms adapt their growth program to changes in energy needs and states, 2) what are the molecular mechanisms that sense nutrient availability and regulate body size, and 3) how do endocrine signals interact with metabolic and growth regulators?
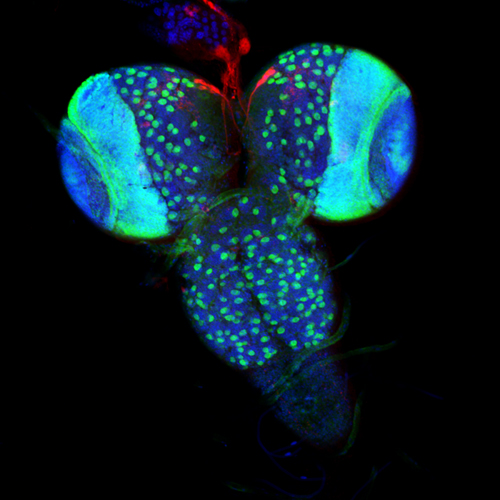
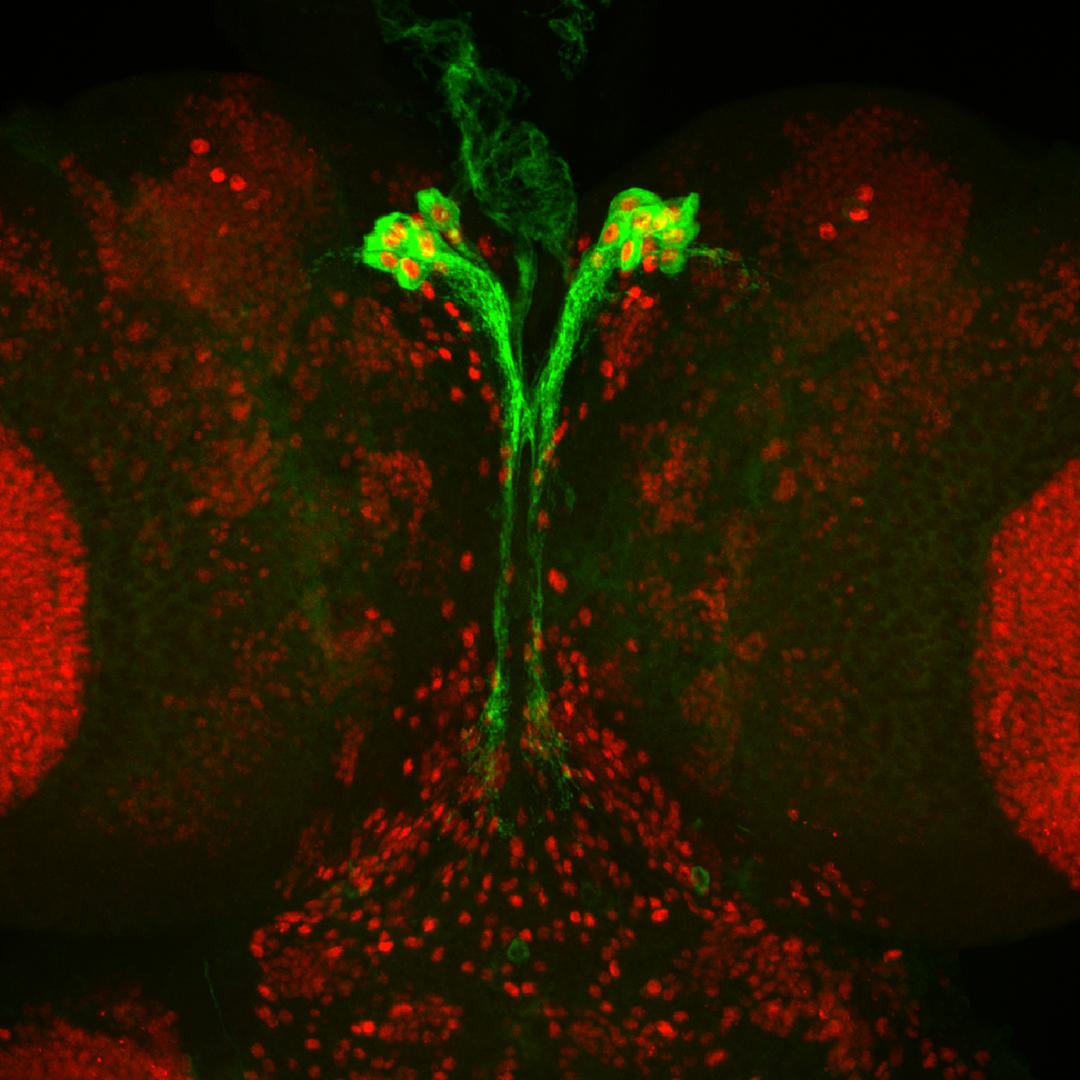
To better understand the interface between nutrient availability and growth regulation, we are focusing on how nutrition controls systemic growth through Drosophila insulin-like peptides (Dilps). Members of the insulin family peptides have conserved roles in the regulation of growth and metabolism in a wide variety of metazoans. We have demonstrated the molecular mechanism underlying the nutrient-dependent expression of a Dilp gene. We have also conducted in vivo RNAi screening to identify new players regulating growth and developmental timing at the organismal level. We described the first demonstration of the glia-derived endocrine factor regulating systemic body growth. Because Dilp regulates both growth and metabolism during development, we are analyzing the physiological significance of the regulation of sugar metabolism by insulin/IGF signaling. Our work focusing on the blood sugar trehalose revealed that metabolism of hemolymph sugar plays a critical role for body growth under poor dietary conditions. Dietary condition-specific phenotype in Drosophila provides new insights into the significance of gene-environment interactions.

Research Theme
- Regulatory mechanisms for cell proliferation during animal development
- Regulatory mechanisms for developmental timing
- Molecular mechanisms for nutrient sensing
Selected Publications
Sasaki A, Nishimura T, Takano T, et al.
white regulates proliferative homeostasis of intestinal stem cells during aging in Drosophila
Nature Metabolism doi: 10.1038/s42255-021-00375-x
Yamada T, Hironaka KI, Habara O, et al.
A developmental checkpoint directs metabolic remodeling as a strategy against starvation in Drosophila.
Nature Metabolism (2020) doi: 10.1038/s42255-020-00293-4
Nishimura T.
Feed-forward regulation of glucose metabolism by steroid hormones drives a developmental transition in Drosophila.
Current Biology 30(18). 3624-3632 E5 (2020) doi: 10.1016/j.cub.2020.06.043
Imura E, Shimada-Niwa Y, Nishimura T, et al.
The Corazonin-PTTH Neuronal Axis Controls Systemic Body Growth by Regulating Basal Ecdysteroid Biosynthesis in Drosophila melanogaster.
Current Biology 30(11). 2156-2165 E5 (2020) doi: 10.1016/j.cub.2020.03.050
Matsushita R, Nishimura T.
Trehalose metabolism confers developmental robustness and stability in Drosophila by regulating glucose homeostasis
Communications Biology 3. 170 (2020) doi: 10.1038/s42003-020-0889-1
Hironaka K, Fujimoto K, Nishimura T.
Optimal scaling of critical size for metamorphosis in the genus Drosophila.
iScience 20. 348-358 (2019) doi: 10.1016/j.isci.2019.09.033
Yasugi T, Yamada T, and Nishimura T.
Adaptation to dietary conditions by trehalose metabolism in Drosophila.
Scientific Reports 7. 1619 (2017) doi :10.1038/s41598-017-01754-9
Yoshida M, Matsuda H, Kubo H, and Nishimura T.
Molecular characterization of Tps1 and Treh genes in Drosophila and their role in body water homeostasis.
Scientific Reports 6. 30582 (2016) doi:10.1038/srep30582
Okamoto N and Nishimura T.
Signaling from glia and cholinergic neurons controls nutrient-dependent production of an insulin-like peptide for Drosophila body growth.
Developmental Cell 35. 295–310 (2015) doi:10.1016/j.devcel.2015.10.003
Matsuda H, Yamada T, Yoshida M, and Nishimura T.
Flies without trehalose.
Journal of Biological Chemistry 290. 1244–1255 (2015) doi:10.1074/jbc.M114.619411
Okamoto N, Nakamori R, Murai T, et al.
A secreted decoy of InR antagonizes insulin/IGF signaling to restrict body growth in Drosophila.
Genes & Development 27. 87–97 (2013) doi:10.1101/gad.204479.112
Okamoto N, Nishimori Y, and Nishimura T.
Conserved role for the Dachshund protein with Drosophila Pax6 homolog Eyeless in insulin expression.
Proceedings of the National Academy of Sciences of the United States of America 109. 2406–2411 (2012) doi:10.1073/pnas.1116050109