Team Director
Kazunari Miyamichi
Ph.D.
Laboratory for Comparative Connectomics
Location Kobe / Developmental Biology Buildings
E-mailkazunari.miyamichi@riken.jp
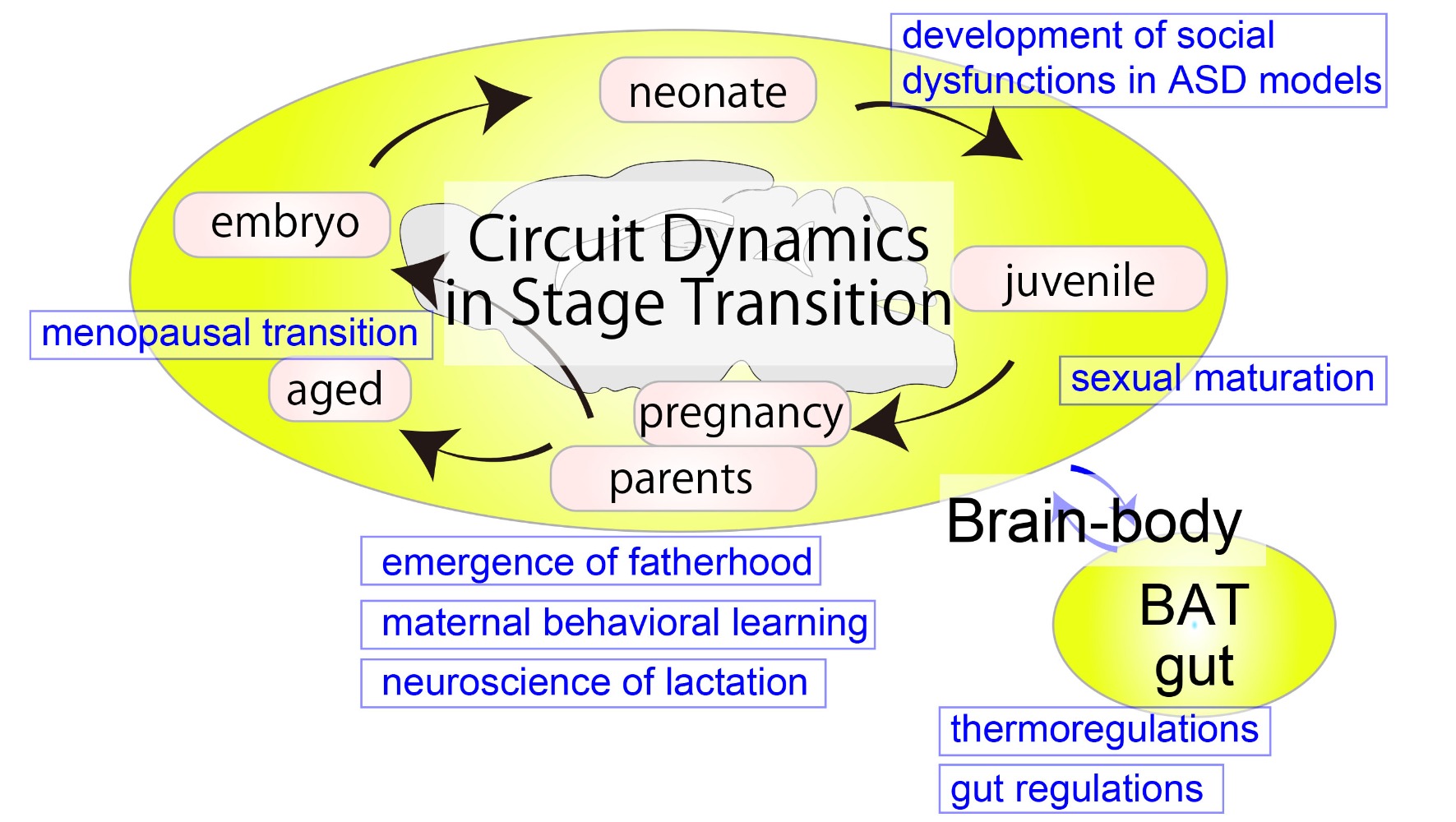
The brain can flexibly adapt the physiology and behaviors of the body to specific life-stage demands. For example, sexual maturation is initiated when the maturation and nutritional state of the body are ready for reproduction. Parental caregiving behaviors to infants are facilitated around the time when their own young are expected. Specific neural activity patterns that regulate milk ejection only emerge in lactating mothers. These examples suggest the presence of neural mechanisms that plastically adjust neural input organizations and output functions based on an internal state of organisms. However, little is known as to how such adaptive plasticity is implemented at the level of neural circuits. Our research aims to elucidate the mechanisms of flexible neuronal functions associated with different life stages in mice by targeting a diverse array of brain regions, ranging from the frontal cortex to the sympathetic nervous system. We employ a combination of cutting-edge techniques, including single-cell RNAseq, virus-based circuit mapping, in vivo imaging of neural activity, and molecular and neural manipulation of specific neuron types. Through our work, we hope to provide novel insights into the plasticity of the nervous system that underlies the control of animal behavior and body physiology.
Research Theme of our laboratory
Research Themes
- Neural basis of behavioral plasticity associated with life-stage transitions
- Neural circuit organizations and functions related to parturition, lactation, and parental caregiving behaviors
- Neural circuit dissection of the sympathetic nervous system for various organs
Selected Publications
Yaguchi K, Miyamichi K, Tasaka GI.
Flexible adjustment of oxytocin neuron activity in mouse dams revealed by microendoscopy.
Science Advances
10(50), eadt1555 (2024)
doi: 10.1126/sciadv.adt1555
Harima Y, Tsurutani M, Yamada S, et al.
Parallel labeled-line organization of sympathetic outflow for selective organ regulation in mice.
Nature Communications
15(1), 10478 (2024)
doi: 10.1038/s41467-024-54928-1
Tsurutani M, Goto T, Hagihara M, et al.
Selective vulnerability of parvocellular oxytocin neurons in social dysfunction.
Nature Communications
15(1), 8661 (2024)
doi: 10.1038/s41467-024-53092-w
Miyamichi K.
Neural basis for behavioral plasticity during the parental life-stage transition in mice.
Frontiers in Neural Circuits
17, 1340497 (2023)
doi: 10.3389/fncir.2023.1340497
Goto T, Hagihara M, Miyamichi K.
Dynamics of pulsatile activities of arcuate kisspeptin neurons in aging female mice.
eLife
12, e82533 (2023)
doi: 10.7554/eLife.82533
Yaguchi K, Hagihara M, Konno A, et al.
Dynamic modulation of pulsatile activities of oxytocin neurons in lactating wild-type mice
PLOS One
18, e0285589 (2023)
doi: 10.1371/journal.pone.0285589
Inada K, Tsujimoto K, Yoshida M, et al.
Oxytocin signaling in the posterior hypothalamus prevents hyperphagic obesity in mice.
eLife
11, e75718 (2022)
doi: 10.7554/eLife.75718
Yukinaga H, Hagihara M, Tsujimoto K, et al.
Recording and manipulation of the maternal oxytocin neural activities in mice.
Current Biology
32(17), 3821-3829 (2022)
doi: 10.1016/j.cub.2022.06.083
Inada K, Hagihara M, Tsujimoto K, et al.
Plasticity of neural connections underlying oxytocin-mediated parental behaviors of male mice.
Neuron
110(12), 2009-2023 (2022)
doi: 10.1016/j.neuron.2022.03.033
Mano T, Murata K, Kon K, et al.
CUBIC-Cloud provides an integrative computational framework toward community-driven whole-mouse-brain mapping.
Cell Reports Methods
1(2), 100038 (2021)
doi: 10.1016/j.crmeth.2021.100038
Yoshihara C, Tokita K, Maruyama T, et al.
Calcitonin receptor signaling in the medial preoptic area enables risk-taking maternal care.
Cell Reports
35(9), 109204 (2021)
doi: 10.1016/j.celrep.2021.109204
Ishii K K, Osakada T, Mori H, et al.
A labeled-line neural circuit for pheromone-mediated sexual behaviors in mice.
Neuron
95, 123-137 (2017)
doi: 10.1016/j.neuron.2017.05.038