
Team Director
Hiroshi Kiyonari
Ph.D.
Laboratory for Animal Resources and Genetic Engineering
Location Kobe / Developmental Biology Buildings
E-mailhiroshi.kiyonari@riken.jp
- Harnessing Experimental Animals -
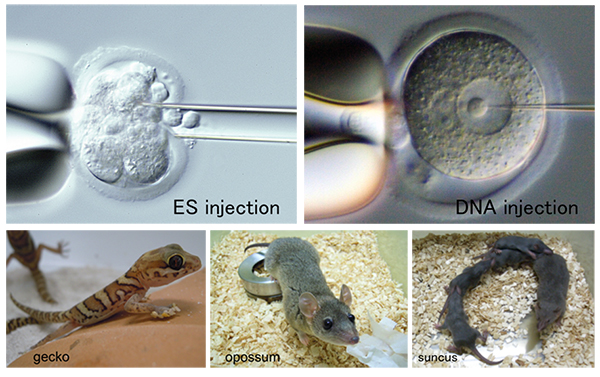
The major mission of the Animal Resource Development Unit is to integrate advanced reproductive and developmental biology technologies and environment for animal research experiments through innovative research in the development of novel experimental animal models and related technical support activities. We utilize our expertise, not only to manage the state-of-the-art SPF rodent facility at RIKEN Kobe Campus, but also to accommodate a variety of animal research needs, including animal material and resource support, technical training, and education for proper conduct of animal research. In recent years, we have established colonies of a Soricomorpha species, suncus (Suncus murinus), a metatherian species, gray short-tail opossum (Monodelphis domestica), and a reptilian species, gecko (Paroedura picta) for prospective distribution services of these new resources to the research community. In addition, in close cooperation with Genetic Engineering Team, we are facilitating joint projects to generate and distribute novel genetically engineered mice with domestic and international biomedical research community, as well as conducting original research projects in live imaging and analyses of early mouse development.
Research Theme
- Advancement of technologies in reproductive biology
- In vitro development of early mouse embryos
Selected Publications
Goto T, Hagihara M, Irie S, et al.
Dietary availability acutely influences puberty onset via a hypothalamic neural circuit.
Neuron
(2025)
doi: 10.1016/j.neuron.2025.01.015
Matsumoto N, Miyano M, Abe T, et al.
Generation of Dopamine Transporter (DAT)-mCherry Knock-in Rats by CRISPR-Cas9 Genome Editing.
Biological & Pharmaceutical Bulletin
47(2), 394-398 (2024)
doi: 10.1248/bpb.b23-00598
Wang Y, Cao S, Tone D, et al.
Postsynaptic competition between calcineurin and PKA regulates mammalian sleep-wake cycles.
Nature
636(8042), 412-421 (2024)
doi: 10.1038/s41586-024-08132-2
Bilgic M, Wu Q, Suetsugu T, et al.
Truncated radial glia as a common precursor in the late corticogenesis of gyrencephalic mammals.
eLife
12, RP91406 (2023)
doi: 10.7554/eLife.91406
Suita K, Ishikawa K, Kaneko M, et al.
Mouse embryonic stem cells embody organismal-level cold resistance.
Cell Reports
42(8), 112954 (2023)
doi: 10.1016/j.celrep.2023.112954
Abe T, Kaneko M, Kiyonari H.
A reverse genetic approach in geckos with the CRISPR/Cas9 system by oocyte microinjection.
Developmental Biology
497, 26-32 (2023)
doi: 10.1016/j.ydbio.2023.02.005
Nishiyama C, Saito Y, Sakaguchi A, et al.
Prolonged Myocardial Regenerative Capacity in Neonatal Opossum.
Circulation
146(2), 125-139 (2022)
doi: 10.1161/CIRCULATIONAHA.121.055269
Kiyonari H, Kaneko M, Abe T, et al.
Targeted gene disruption in a marsupial, Monodelphis domestica, by CRISPR/Cas9 genome editing.
Current Biology
31(17), 3956-3963 (2021)
doi: 10.1016/j.cub.2021.06.056
Abe T, Inoue KI, Furuta Y, Kiyonari H.
Pronuclear Microinjection during S-Phase Increases the Efficiency of CRISPR-Cas9-Assisted Knockin of Large DNA Donors in Mouse Zygotes.
Cell Reports
31(7), 107653 (2020)
doi: 10.1016/j.celrep.2020.107653
Kiyonari H, Kaneko M, Abe T, et al.
Dynamic organelle localization and cytoskeletal reorganization during preimplantation mouse embryo development revealed by live imaging of genetically encoded fluorescent fusion proteins.
Genesis.
57(2), e23277 (2019)
doi: 10.1002/dvg.23277
Members
Hiroshi Kiyonari
Team Director
Takaya Abe
Technical Scientist
Kenichi Inoue
Expert Technician
Mayo Shigeta
Expert Technician
Miho Sato
Technical Staff
Naoko Shimada
Technical Staff
Aki Shiraishi
Technical Staff
Hinako Takase
Technical Staff
Tomoko Tokuhara
Technical Staff
Yuka Nakatani
Technical Staff
Miwako Nomura
Technical Staff
Junko Hara
Technical Staff
Kana Bando
Technical Staff
Michiko Higashikawa
Technical Staff
Yoshino Matsumoto
Technical Staff
Anna Yasunaga
Technical Staff
Daisuke Yamamoto
Technical Staff
Riko Yoshimi
Technical Staff
Megumi Watase
Technical Staff
Xuchi Pan
Research Associate
Masayo Okugawa
Assistant
Mika Hikawa
Assistant





