
Team Director
Tomoya Kitajima
Ph.D.
Laboratory for Chromosome Segregation
Location Kobe / Developmental Biology Buildings
E-mail tomoya.kitajima[at]riken.jp
Please replace [at] with @.
The oocyte becomes an egg through meiosis. The egg fertilizes with a sperm and undergoes repeated cell divisions to give rise to an entire body. We study chromosome segregation during meiosis in oocytes and during mitosis in fertilized eggs, taking advantage of techniques for high-throughput and high-resolution live imaging of mouse oocytes combined with micromanipulation and genetic engineering methods. The first cell division that oocytes undergo is meiosis I. Chromosome segregation in this division is error-prone and the rate of errors increases with maternal age. Subsequently, chromosomes are segregated in meiosis II upon fertilization, and then segregated again in mitosis after DNA replication. We will reveal distinct mechanisms for chromosome segregation during these subsequent but fundamentally different cell divisions. By uncovering the mechanism of chromosome segregation during meiosis I in oocytes, we understand why oocyte meiosis I is error-prone and related to age. Comparing the mechanisms in meiosis I with those found in meiosis II and mitosis may provide insights into the capacity of cells to flexibly use different strategies for chromosome segregation. The findings will be exploited to collaborative studies with reproductive medicine.
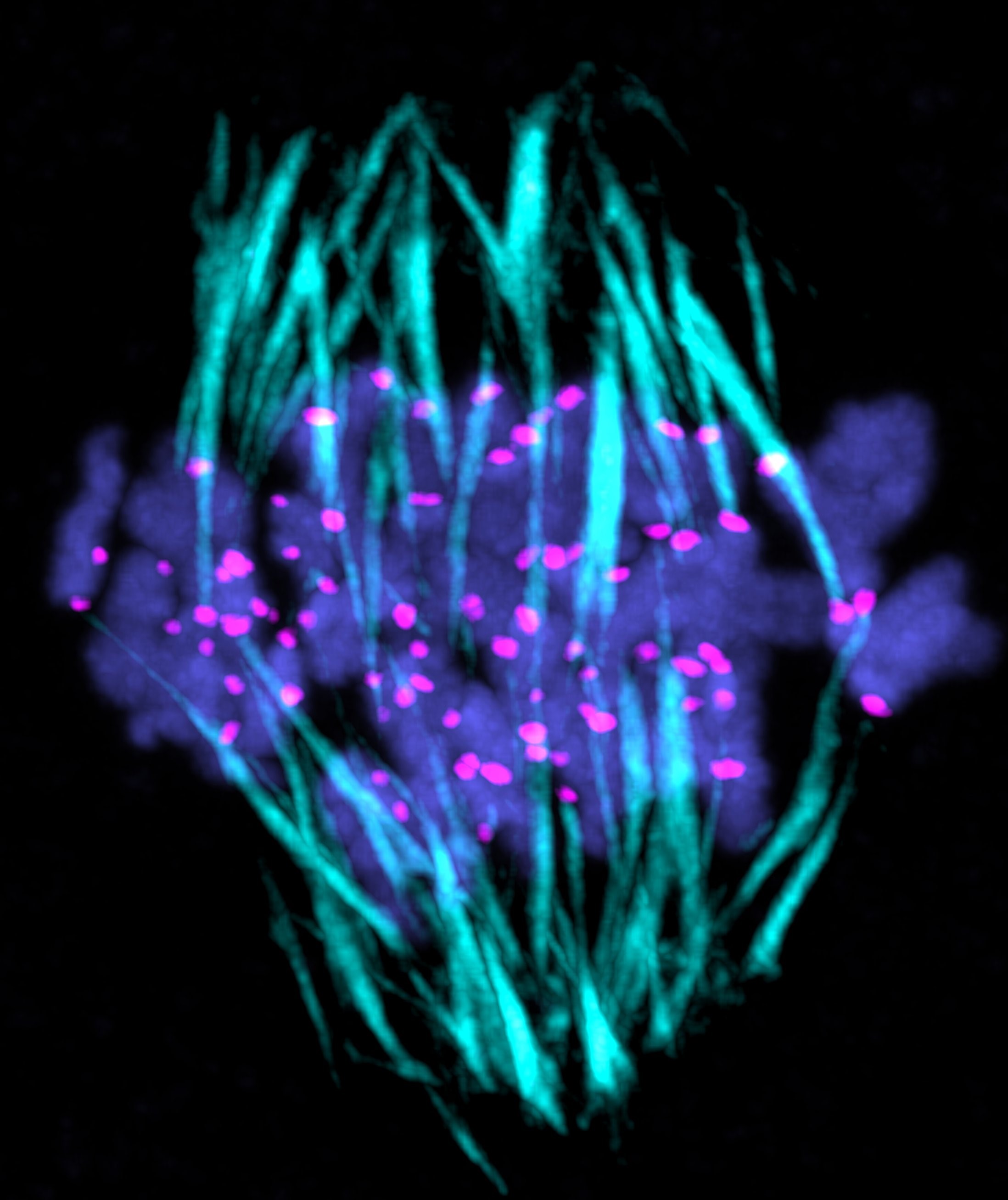
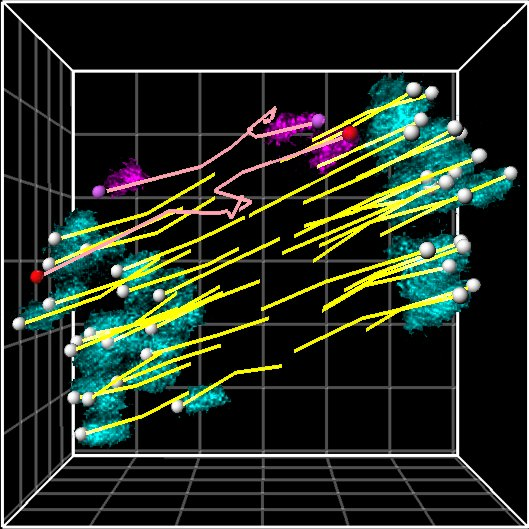
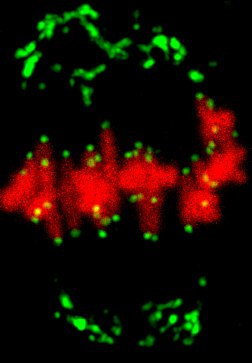
Research Theme
- Analysis of the mechanisms underlying meiotic chromosome segregation in mammalian oocytes
- Study of the mechanisms underlying chromosome segregation during mitosis in fertilized eggs
- Age-related errors in oocytes and fertilized eggs
Selected Publications
Asai K, Zhou Y, Takenouchi O, et al.
Artificial kinetochore beads establish a biorientation-like state in the spindle.
Science
385(6715), 1366-1375 (2024)
doi: 10.1126/science.adn5428
Takahashi S, Kyogoku H, Hayakawa T, et al.
Embryonic genome instability upon DNA replication timing program emergence.
Nature
633, 686-694 (2024)
doi: 10.1038/s41586-024-07841-y
Takenouchi O, Sakakibara Y, Kitajima TS.
Live chromosome identifying and tracking reveals size-based spatial pathway of meiotic errors in oocytes.
Science
385(6706), (2024)
doi: 10.1126/science.adn5529
Mishina T, Tabata N, Hayashi T, et al.
Single-oocyte transcriptome analysis reveals aging-associated effects influenced by life stage and calorie restriction.
Aging Cell
20(8), e13428 (2021)
doi: 10.1111/acel.13428
Courtois A, Yoshida S, Takenouchi O, et al.
Stable kinetochore-microtubule attachments restrict MTOC position and spindle elongation in oocytes.
EMBO Reports
22(4), e51400 (2021)
doi: 10.15252/embr.202051400
Yoshida S, Nishiyama S, Lister L, et al.
Prc1-rich kinetochores are required for error-free acentrosomal spindle bipolarization during meiosis I in mouse oocytes.
Nature Communications
11, 2652 (2020)
doi: 10.1038/s41467-020-16488-y
Ding Y, Kaido M, Llano E, et al.
The post-anaphase SUMO pathway ensures the maintenance of centromeric cohesion through meiosis I-II transition in mammalian oocytes.
Current Biology
28(10), 1661-1669 (2018)
doi: 10.1016/j.cub.2018.04.019
Kyogoku H, Kitajima TS.
Large cytoplasm is linked to the error-prone nature of oocytes.
Developmental Cell
41(3), 287-298 (2017)
doi: 10.1016/j.devcel.2017.04.009
Sakakibara Y, Hashimoto S, Nakaoka Y, et al.
Bivalent separation into univalents precedes age-related meiosis I errors in oocytes.
Nature Communications
6, 7550 (2015)
doi: 10.1038/ncomms8550
Yoshida S, Kaito M, Kitajima TS.
Inherent instability of correct kinetochore-microtubule attachments during meiosis I in oocytes.
Developmental Cell
33(5), 589-602 (2015)
doi: 10.1016/j.devcel.2015.04.020
Members
Tomoya Kitajima
Team Director
Shuhei Yoshida
Technical Scientist
So Shimamoto
Special Postdoctoral Researcher
Manami Koshiguchi
Postdoctoral Researcher
Mihoko Fushii
Postdoctoral Researcher
Hirohisa Kyogoku
Visiting Scientist
Kaori Hamada
Technical Staff II
Kohei Asai
Research Part-time Worker I
Yuanzhuo Zhou
Junior Research Associate
MeiAkiko Mukose
Junior Research Associate
Remi Kanemura
Junior Research Associate
Miho Sakuma
Student Trainee
Emiko Ichihara
Student Trainee
Angela Jennifer Tantry
Student Trainee
News

Nov. 11, 2025 BDR News
BDR Team Director Tomoya Kitajima awarded the Osaka Science Prize

Sep. 11, 2025 Research
How egg cells control the timing of cell division
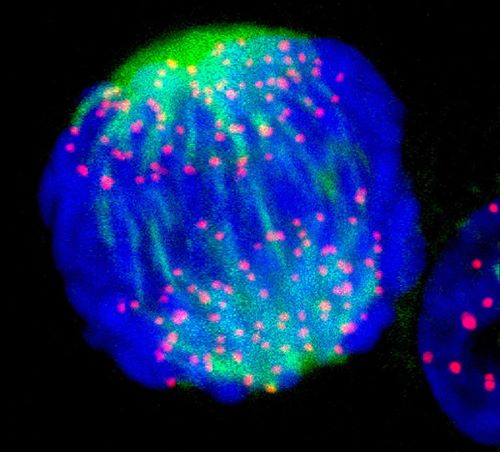
Jan. 9, 2025 Research
Synthetic beads mimic critical process in cell division, opening new paths for biomachines
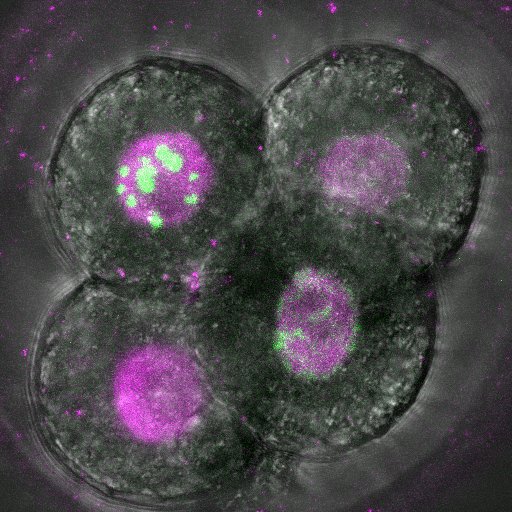
Dec. 24, 2024 Research
DNA copying hits a snag in early embryos
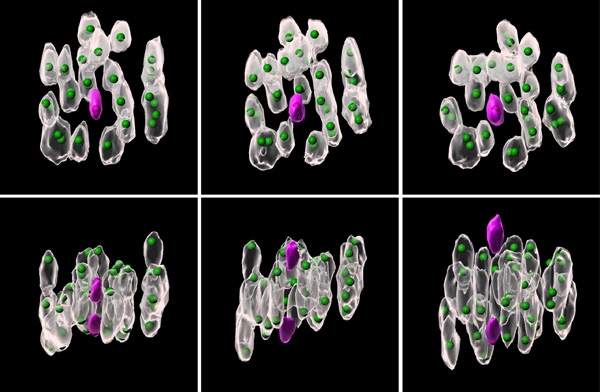
Oct. 10, 2024 Research
Unlocking the mystery of chromosomal errors

Oct. 20, 2023 Research
Stolen genes allow parasitic control of behavior
Sep. 12, 2022 Research
Smaller eggs enhance IVF outcomes for male infertility in mice
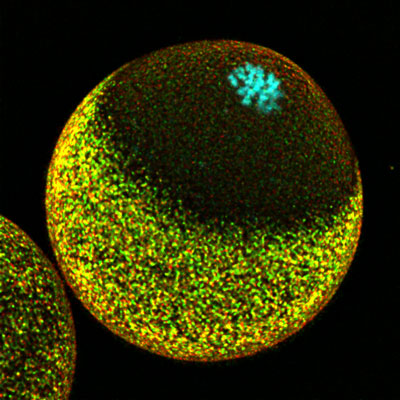
Nov. 26, 2021 Research
Successful fertilization requires careful coordination of chromosomes

Dec. 17, 2020 Research
Oh so simple: Eight genes enough to convert mouse stem cells into oocyte-like cells
Jul. 31, 2020 Research
Mice need kinetochores rich in a microtubule crosslinker to achieve error-free oocyte division
Jul. 20, 2018 Research
Mechanism identified that stabilizes chromosome pairs during meiosis




