
Unit Leader
Miki Ebisuya
Ph.D.
[Closed Mar. 2019]
In our lab, we strive to create or reconstitute multicellular developmental mechanisms. Our aim of reconstitution is to test the sufficiency of current understandings of mechanisms of interest, as well as to discover unexplained or unexpected elements through observation. The fundamental principles of development include cell autonomous differentiation, spatio-temporal pattern formation and tissue deformation. Thus, we have been trying to reconstitute these principles one by one and so far succeeded in reconstituting cell autonomous differentiation. Namely, we created an artificial gene circuit mimicking Delta-Notch lateral inhibition in mammalian cell culture, causing spontaneous bifurcation of initially identical cells into two different cell types (Matsuda et al, Nat Commun 2015). Now we are working on the reconstitution of reaction-diffusion pattern formation, intercellular synchronized oscillation and 3D-tissue deformation. Artificially reconstituted systems also have the advantage of facilitating measurements and the modification of parameters, which we hope will contribute to the quantitative understanding of developmental principles.
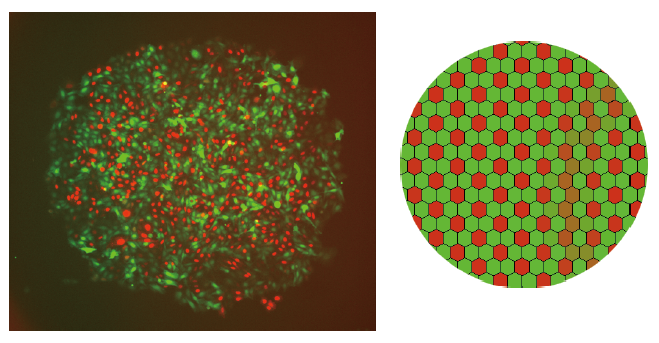
A cell colony showing a salt-and-pepper pattern of red cells and green cells (Left: Cells engineered with a synthetic gene circuit, Right: Simulation)
Research Theme
- Reconstitution of cell autonomous differentiation
- Reconstitution of cell pattern formation
- Reconstitution of synchronized oscillation
- Reconstitution of tissue deformation



