Team Director
Shun-ichi Sekine
Ph.D.
Laboratory for Transcription Structural Biology
[Affiliation has changed to RIKEN Center for Integrated Medical Sciences (IMS) as of April 2025]
LocationYokohama
E-mailshunichi.sekine@riken.jp
Life activities are supported by numerous biomolecules, which communicate with each other and constitute huge assemblies to fulfill diverse cellular needs. In order to visualize their architecture and elucidate their action mechanisms, our team analyzes their 3D structures by cryo-electron microscopy and X-ray crystallography. Our main focus is on huge complexes such as complexes of RNA polymerase and transcription factors, and we unravel molecular mechanisms behind various important biological phenomena. Furthermore, viral or bacterial proteins involved in replication and/or transcription are one of the major drug targets, and our team provides the structural foundations for the drug discovery research.
Research Theme
- Structural bases of transcription and its-related phenomena
- Advancement of technologies for the structural analyses of supramolecular complexes
- Structural basis of viral and bacterial replication/transcription toward drug discovery
Selected Publications
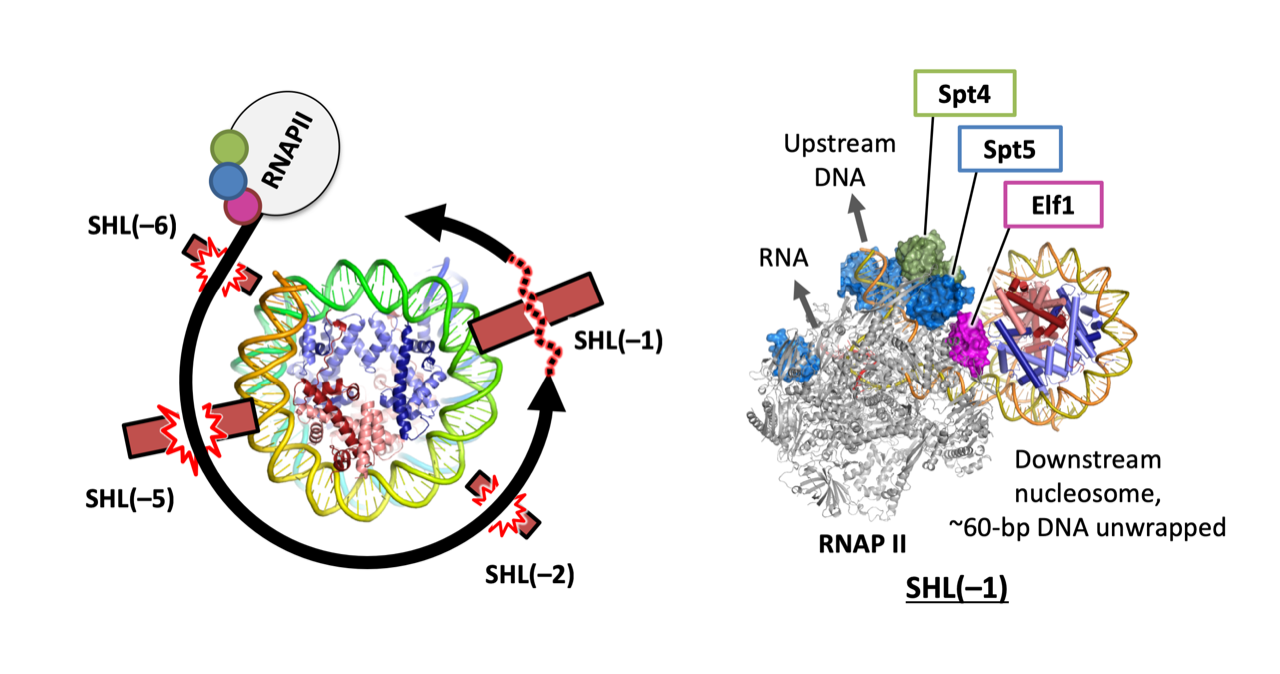
Naganuma M, Kujirai T, Ehara H, et al.
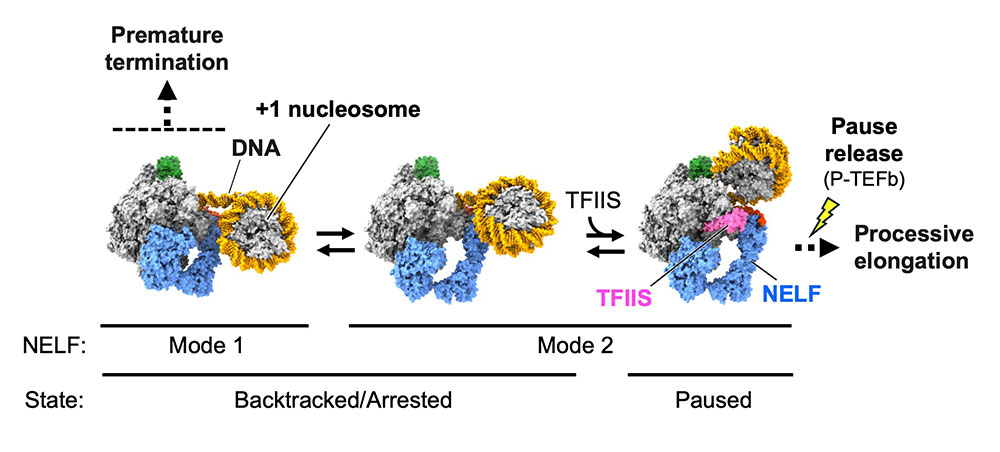
Structural insights into promoter-proximal pausing of RNA polymerase II at +1 nucleosome.
Science Advances
11(10), eadu0577 (2025)
doi: 10.1126/sciadv.adu0577
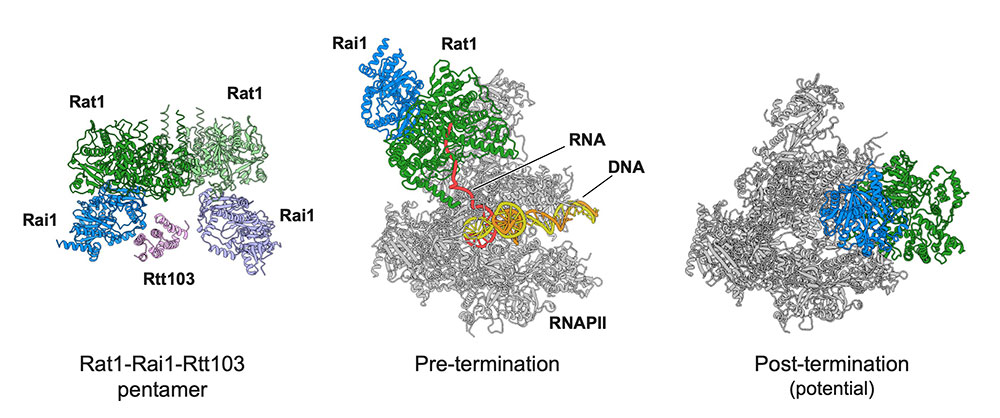
Yanagisawa T, Murayama Y, Ehara H, et al.
Structural basis of eukaryotic transcription termination by the Rat1 exonuclease complex
Nature Communications
15, 7854 (2024)
doi: 10.1038/s41467-024-52157-0
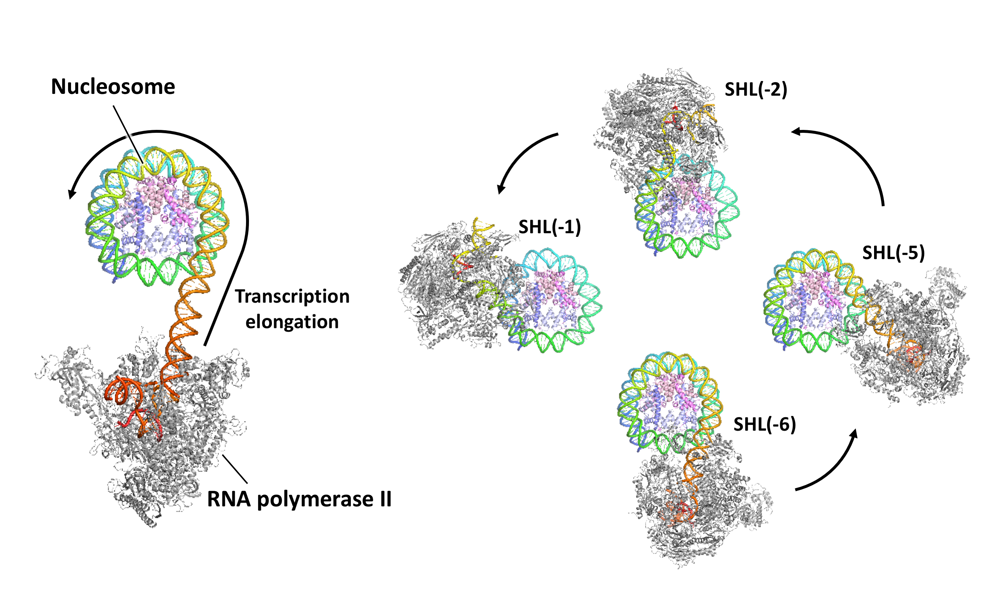
Akatsu M, Ehara H, Kujirai T, et al.
Cryo-EM structures of RNA polymerase II-nucleosome complexes rewrapping transcribed DNA
J Biol Chem
299(12), 105477 (2023)
doi: 10.1016/j.jbc.2023.105477
Sekine S, Ehara H, Kujirai T, et al.
Structural perspectives on transcription in chromatin
Trends in Cell Biology
(2023)
doi: 10.1016/j.tcb.2023.07.011
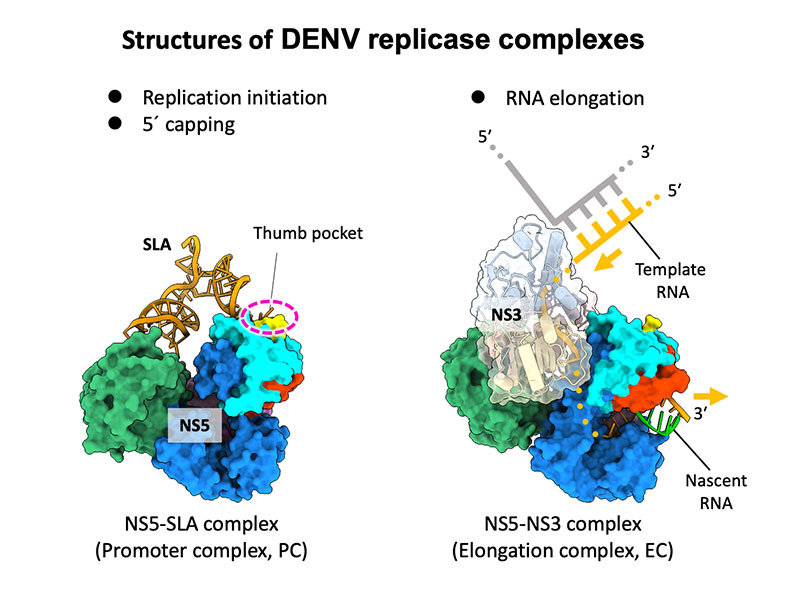
Osawa T, Aoki M, Ehara H, et al.
Structures of dengue virus RNA replicase complexes
Molecular Cell
(2023)
doi: 10.1016/j.molcel.2023.06.023
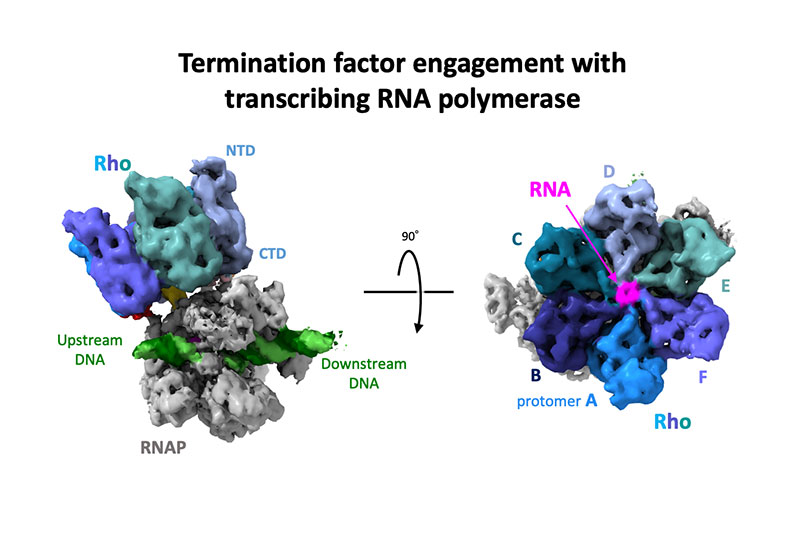
Murayama Y, Ehara H, Aoki M, et al.
Structural basis of the transcription termination factor Rho engagement with transcribing RNA polymerase from Thermus thermophilus
Science Advances
9(6), eade7093 (2023)
doi: 10.1126/sciadv.ade7093
Hirano R, Ehara H, Kujirai T, et al.
Structural basis of RNA polymerase II transcription on the chromatosome containing linker histone H1
Nature Communications
13, 7287 (2022)
doi: 10.1038/s41467-022-35003-z
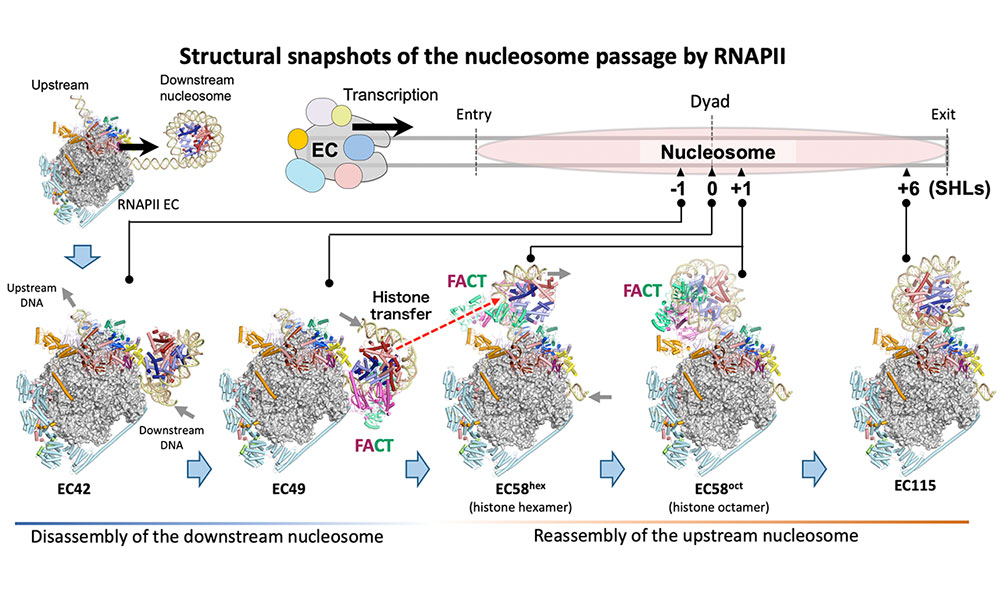
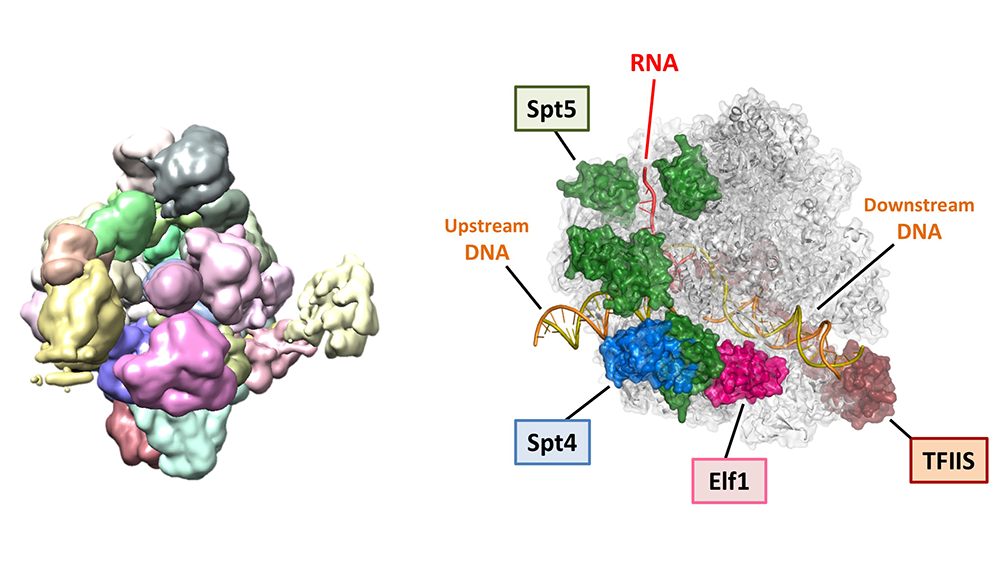
Ehara H, Kujirai T, et al.
Structural basis of nucleosome disassembly and reassembly by RNAPII elongation complex with FACT
Science
377(6611), abp9466 (2022)
doi: 10.1126/science.abp9466
Ehara H, Kujirai T, et al.
Structural insight into nucleosome transcription by RNA polymerase II with elongation factors.
Science
363(6428), 744-747 (2019)
doi: 10.1126/science.aav8912
Shimizu H, Saito A, Mikuni J, et al.
Discovery of a small molecule inhibitor targeting dengue virus NS5 RNA-dependent RNA polymerase.
Plos Neglected Tropical Diseases
13(11), e0007894 (2019)
doi: 10.1371/journal.pntd.0007894
Kujirai T, Ehara H, et al.
Structural basis of the nucleosome transition during RNA polymerase II passage.
Science
362(6414), 595-598 (2018)
doi: 10.1126/science.aau9904
Ooi WY, Murayama Y, Mekler V, et al.
A Thermus phage protein inhibits host RNA polymerase by preventing template DNA strand loading during open promoter complex formation.
Nucleic Acids Research
46(1), 431-441 (2017)
doi: 10.1093/nar/gkx1162
Ehara H, Yokoyama T, Shigematsu H, et al.
Structure of the complete elongation complex of RNA polymerase II with basal factors.
Science
357(6354), 921-924 (2017)
doi: 10.1126/science.aan8552
Sekine S, Murayama Y, Svetlov V, et al.
The ratcheted and ratchetable structural states of RNA polymerase underlie multiple transcriptional functions.
Molecular Cell
57(3), 408-421 (2015)
doi: 10.1016/j.molcel.2014.12.014
Members

Team DirectorShun-ichi Sekine
- shunichi.sekine@riken.jp
- CV
News

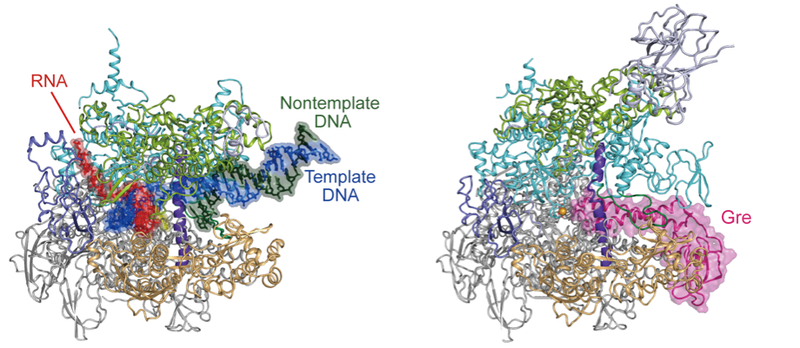
Aug. 12, 2025 Research
Pressing pause on DNA transcription
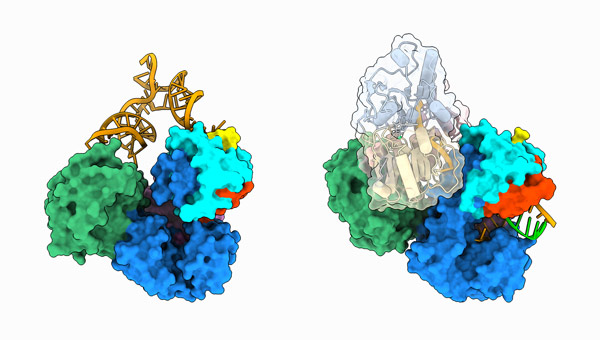
Oct. 31, 2023 Research
Atomic picture of dengue replication could transform antiviral approaches
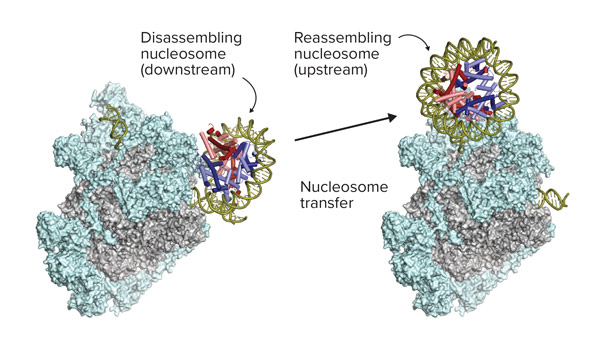
Nov. 17, 2022 Research
Gene-reading enzyme razes and rebuilds DNA-winding structures in its path

Apr. 28, 2022 BDR News
Four BDR scientists awarded MEXT prizes
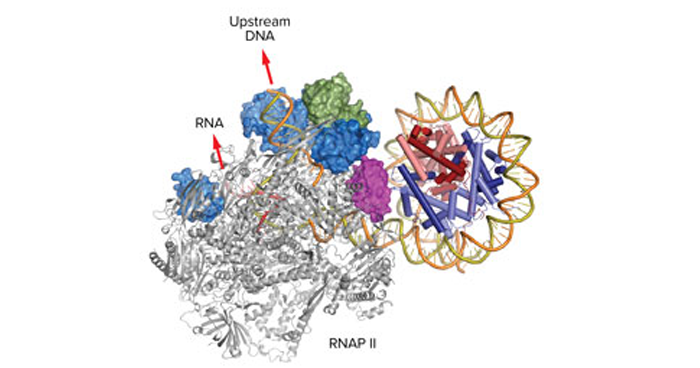
Apr. 12, 2019 Research
Elongation factors smooth transcription in the nucleosome