
Team Leader*
Takanori Kigawa
D. Sci.
Laboratory for Cellular Structural Biology
[Closed Mar. 2023]
E-mailkigawa[at]riken.jp
Please replace [at] with @.
*Current position: Senior Scientist, NMR Operation Team;
Deputy Team Leader, Laboratory for Dynamic Structure of Biomolecules
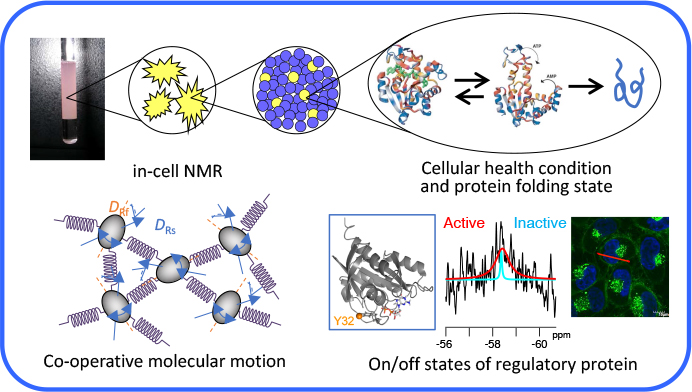
In actual biological cellular environments, biomolecules such as proteins are working dynamically and co-operatively, generally surrounded by high concentrations of macromolecules, so-called “macromolecular crowding” and also “confined” by/in the plasma membrane and/or cellular organelles. In recent years, it has become important to directly investigate the structure and the dynamics of biomolecules in cellular environments because protein behaviors in cells are, at least in some cases, different from those in a dilute, homogeneous solution. We will elucidate cellular events at atomic resolution by analyzing structural dynamics of biomolecules in the cellular environment mainly by using NMR spectroscopy integrated with information science technologies. We will also develop and improve the technologies for sample preparation, stable-isotope labeling, NMR measurements, and data analyses in order to address the issues of low sensitivity and resolution from which NMR measurement of biomolecules in cellular environments usually suffers. In addition, based on our findings, we will develop new technologies by taking advantage of biological functions, for example, the development of technologies for bioelectricity generation based on the mechanisms of energy generation in the living cells.
Project
- NMR analysis of biomolecular structural dynamics in cellular environments
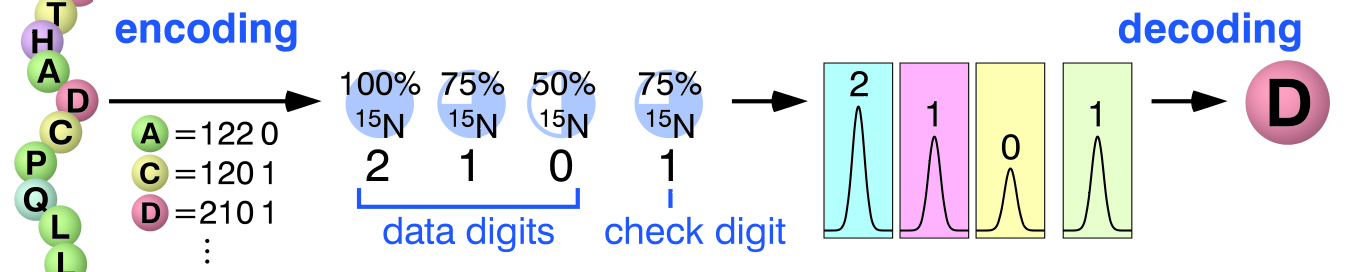
- Development of NMR-related technologies with information sciences
- Development of technologies for stable-isotope labeling of proteins using cell-free synthesis
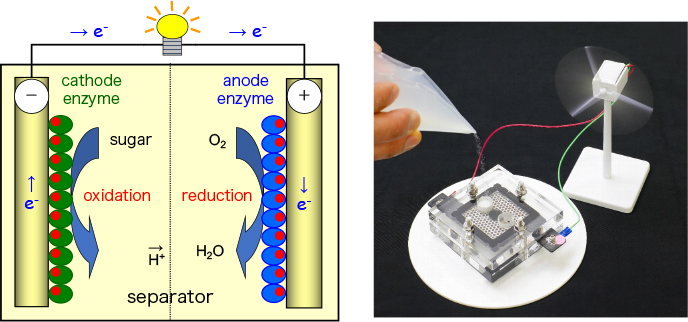
- Development of technology for bioelectricity generation by taking advantage of biological functions
Selected Publications
Yagi H, Kasai T, Rioual E, et al.
Molecular mechanism of glycolytic flux control intrinsic to human phosphoglycerate kinase.
Proceedings of the National Academy of Sciences of the United States of America
118(50), e2112986118 (2021)
doi: 10.1073/pnas.2112986118
Shitanda I, Morigayama Y, Iwashita R, et al.
Paper-based lactate biofuel cell array with high power output.
Journal of Power Sources
489, 229533 (2021)
doi: 10.1016/j.jpowsour.2021.229533
Ito K, Murayama Y, Kurokawa Y, et al.
Real-time tracking reveals catalytic roles for the two DNA binding sites of Rad51.
Nature Communications
11, 2950 (2020)
doi: 10.1038/s41467-020-16750-3
Higuchi K, Yabuki T, Ito M, Kigawa T.
Cold shock proteins improve E. coli cell-free synthesis in terms of soluble yields of aggregation-prone proteins.
Biotechnology and Bioengineering
117(6), 1628-1639 (2020)
doi: 10.1002/bit.27326
Kasai T, Ono S, Koshiba S, et al.
Amino-acid selective isotope labeling enables simultaneous overlapping signal decomposition and information extraction from NMR spectra.
Journal of Biomolecular NMR
74, 125-137 (2020)
doi: 10.1007/s10858-019-00295-9
Inomata K, Kamoshida H, Ikari M, et al.
Impact of cellular health conditions on the protein folding state in mammalian cells.
Chemical Communications (Cambridge)
53(81), 11245-11248 (2017)
doi: 10.1039/c7cc06004a
Kigawa T.
Advances in stable isotope assisted labeling strategies with information science.
Archives of Biochemistry and Biophysics
628, 17-23 (2017)
doi: 10.1016/j.abb.2017.06.014
Kasai T, Nagata K, Okada M, Kigawa T.
NMR spectral analysis using prior knowledge.
Journal of Physics: Conference Series
699(1), 012003 (2016)
doi: 10.1088/1742-6596/699/1/012003
Okamura H, Nishimura H, Nagata T, et al.
Accurate and molecular-size-tolerant NMR quantitation of diverse components in solution.
Scientific Reports
6, 21742 (2016)
doi: 10.1038/srep21742
Shigeno-Nakazawa Y, Kasai T, Ki S, et al.
A pre-metazoan origin of the CRK gene family and co-opted signaling network.
Scientific Reports
6, 34349 (2016)
doi: 10.1038/srep34349
Kasai T, Koshiba S, Yokoyama J, Kigawa T.
Stable isotope labeling strategy based on coding theory.
Journal of Biomolecular NMR
63(2), 213-221 (2015)
doi: 10.1007/s10858-015-9978-8
Harada R, Tochio N, Kigawa T, et al.
Reduced native state stability in crowded cellular environment due to protein-protein interactions.
Journal of the American Chemical Society
135(9), 3696-3701 (2013)
doi: 10.1021/ja3126992
Matsuda T, Furumoto S, Higuchi K, et al.
Rapid biochemical synthesis of C-11-labeled single chain variable fragment antibody for immuno-PET by cell-free protein synthesis.
Bioorganic & Medicinal Chemistry
20(22), 6579-6582 (2012)
doi: 10.1016/j.bmc.2012.09.038
Akama S, Yamamura M, Kigawa T.
A Multiphysics Model of In Vitro Transcription Coupling Enzymatic Reaction and Precipitation Formation.
Biophysical Journal
102(2), 221-230 (2012)
doi: 10.1016/j.bpj.2011.12.014
Yokoyama J, Matsuda T, Koshiba S, et al.
A practical method for cell-free protein synthesis to avoid stable isotope scrambling and dilution.
Analytical Biochemistry
411(2), 223-229 (2011)
doi: 10.1016/j.ab.2011.01.017




