Team Leader*
Takashi Umehara
Ph.D.
Laboratory for Epigenetics Drug Discovery
[Closed Mar. 2023]
E-mailtakashi.umehara[at]riken.jp
Please replace [at] with @.
*Current position: Senior Scientist, Drug Discovery Structural Biology Platform Unit
Epigenetics is a research field that tackles life information that is "added" to genetic information. The major entity of this "addition" is a chemical modification to a complex of a genomic DNA and histone proteins called chromatin. Since epigenetics is involved in the control of cancer, iPS cells and lifespan, it attracts attention as a molecular target for drug discovery, regenerative medicine, and anti-aging. Our laboratory aims to manipulate diseases, iPS cells and anti-aging as desired by developing technologies to precisely reconstitute, detect and control human epigenetic information.
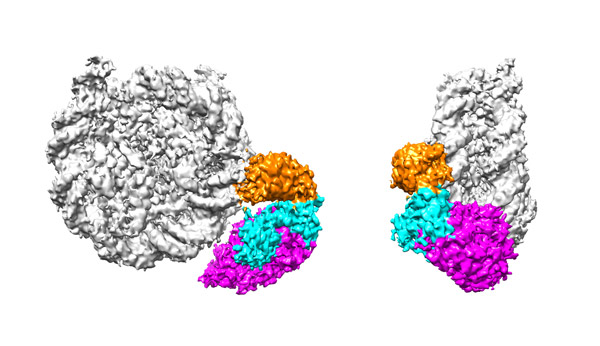
Reconstitution of epigenetics
Synthetic technologies of a protein acetylated as designed. With this technology, transcriptionally active chromatin can be precisely reproduced in vitro.
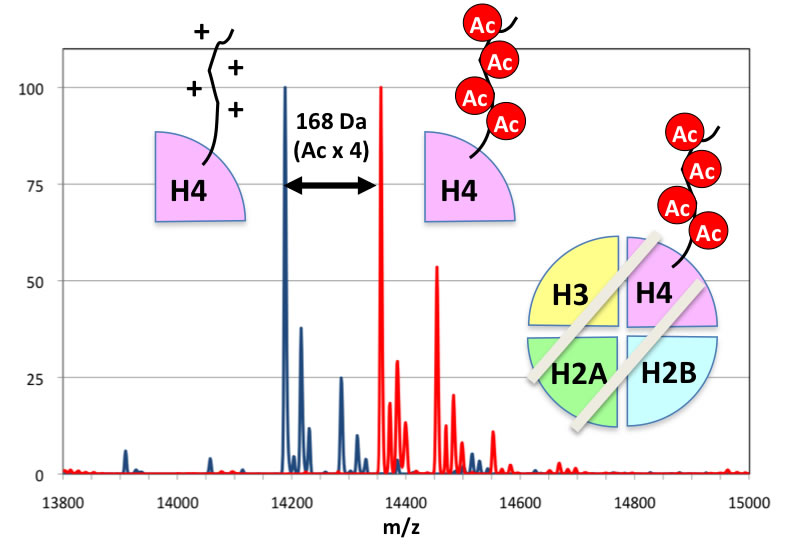
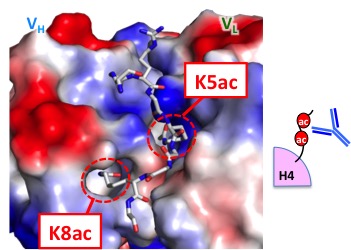
Detection of epigenetics
Development of an antibody that detects acetylation at two residues in histone H4 tail. With this antibody, transcriptionally hyperactive chromatin can be detected at single nucleosome resolution.
Regulation of epigenetics
Histone demethylase inhibitor S2101 developed based on protein structure. This compound is commercially available as an epigenetics-regulating reagent.

Selected Publications
Kikuchi M, Takase S, Konuma T, et al.
GAS41 promotes H2A.Z deposition through recognition of the N terminus of histone H3 by the YEATS domain.
Proceedings of the National Academy of Sciences of the United States of America
120(43), e2304103120 (2023)
doi: 10.1073/pnas.2304103120
Das ND, Chang JC, Hon CC, et al.
Defining super-enhancers by highly ranked histone H4 multi-acetylation levels identifies transcription factors associated with glioblastoma stem-like properties.
BMC Genomics
24(1), 574 (2023)
doi: 10.1186/s12864-023-09659-w
Kikuchi M, Morita S, Wakamori M, et al.
Epigenetic mechanisms to propagate histone acetylation by p300/CBP.
Nature Communications
14(1), 4103 (2023)
doi: 10.1038/s41467-023-39735-4
Liu N, Konuma T, Sharma R, et al.
Histone H3 lysine 27 crotonylation mediates gene transcriptional repression in chromatin.
Molecular Cell
83(13), 2206-2221 (2023)
doi: 10.1016/j.molcel.2023.05.022
Noritsugu K, Suzuki T, Dodo K, et al.
Lysine long-chain fatty acylation regulates the TEAD transcription factor.
Cell Reports
42(4), 112388 (2023)
doi: 10.1016/j.celrep.2023.112388
Takase S, Hiroyama T, Shirai F, et al.
A specific G9a inhibitor unveils BGLT3 lncRNA as a universal mediator of chemically induced fetal globin gene expression.
Nature Communications
14(1), 23 (2023)
doi: 10.1038/s41467-022-35404-0
Niwa H, Watanabe C, Sato S, et al.
Structure–activity relationship and in silico evaluation of cis- and trans-PCPA-derived inhibitors of LSD1 and LSD2.
ACS Medicinal Chemistry Letters
13, 1485-1492 (2022)
doi: 10.1021/acsmedchemlett.2c00294
Kikuchi M, Morita S, Goto M, et al.
Elucidation of binding preferences of YEATS domains to site-specific acetylated nucleosome core particles.
Journal of Biological Chemistry
298, 102164 (2022)
doi: 10.1016/j.jbc.2022.102164
Sasaki K, Suzuki M, Sonoda T, et al.
Visualization of the dynamic interaction between nucleosomal histone H3K9 tri-methylation and HP1α chromodomain in living cells.
Cell Chemical Biology
29, 1153-1161 (2022)
doi: 10.1016/j.chembiol.2022.05.006
Koda Y, Sato S, Yamamoto H, et al.
Design and synthesis of tranylcypromine-derived LSD1 inhibitors with improved hERG and microsomal stability profiles.
ACS Medicinal Chemistry Letters
13, 848-854 (2022)
doi: 10.1021/acsmedchemlett.2c00120
Kitagawa H, Kikuchi M, Sato S, et al.
Structure-based identification of potent lysine-specific demethylase 1 inhibitor peptides and temporary cyclization to enhance proteolytic stability and cell growth-inhibitory activity.
Journal of Medicinal Chemistry
64, 3707-3719 (2021)
doi: 10.1021/acs.jmedchem.0c01371
Wakamori M, Okabe K, Ura K, et al.
Quantification of the effect of site-specific histone acetylation on chromatin transcription rate.
Nucleic Acids Research
48(22), 12648-12659 (2020)
doi: 10.1093/nar/gkaa1050
Wu HD, Kikuchi M, Dagliyan O, et al.
Rational design and implementation of a chemically inducible hetero-trimerization system.
Nature Methods
17, 928-936 (2020)
doi: 10.1038/s41592-020-0913-x
Furukawa A, Wakamori M, Arimura Y, et al.
Acetylated histone H4 tail enhances histone H3 tail acetylation by altering their mutual dynamics in the nucleosome.
Proceedings of the National Academy of Sciences USA
117, 19661-19663 (2020)
doi: 10.1073/pnas.2010506117
Saito S, Kikuchi J, Koyama D, et al.
Eradication of central nervous system leukemia of T-cell origin with a brain-permeable LSD1 inhibitor.
Clinical Cancer Research
25, 1601-1611 (2019)
doi: 10.1158/1078-0432.CCR-18-0919
Handoko L, Kaczkowski B, Hon CC, et al.
JQ1 affects BRD2-dependent and independent transcription regulation without disrupting H4-hyperacetylated chromatin states.
Epigenetics
13, 410-431 (2018)
doi: 10.1080/15592294.2018.1469891