Team Leader
Kensaku Sakamoto
Ph.D.
Laboratory for Nonnatural Amino Acid Technology
[Closed Mar. 2023]
E-mailkensaku.sakamoto[at]riken.jp
Please replace [at] with @.
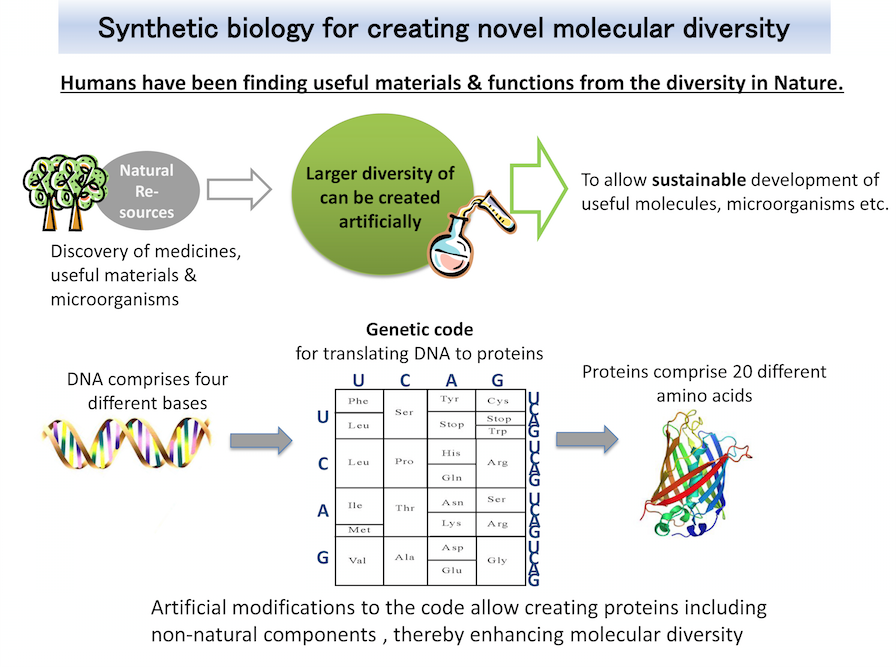
What is Synthetic biology?
Proteins useful for life-science studies and medical applications can be synthesized in living cells programmed with the blueprint for the proteins. The cells engineered to utilize novel amino acids in proteins will facilitate the addition of artificial marks onto proteins, and also the synthesis of proteins linked to pharmaceuticals. These cells will help to realize new structures and functions of proteins. We also exploit this platform technology to provide unique, technical supports for drug discovery.
Collaborations with Structural Biology
Synthetic Biology should work together with Structural Biology, to develop novel proteins and enzymes based on structural knowledge. Not only for engineering, has such collaboration proved useful for elucidating structural basis for the functions of proteins, receptors, and enzymes.
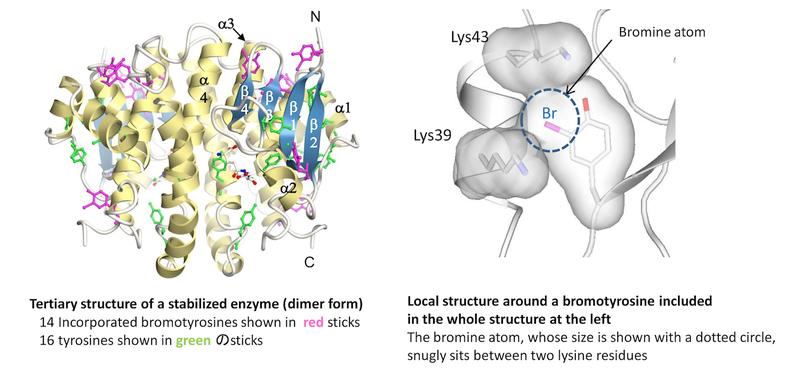
Novel protein stabilization mechanism: Proteins can structurally be stabilized by incorporating non-natural amino acids (bromotyrosines in this case) into them. The left panel shows the tertiary structure of a stabilized enzyme, which was revealed by X-ray crystallography. The right panel shows the local structure around a bromotyrosine included in the whole structure at the left. This amino acids is snugly located between two lysine residues, thereby contributing to the enhanced stability of the enzyme. The other incorporated bromotyrosines also contribute to the stability in similar manners.
Research Theme
- Engineering the genetic code in living cells
- Developing novel proteins with non-natural amino acids
- Technology development to engineer antibodies useful as biologics
Selected Publications
Wada A, Umeki Y, Annoura T, Saito-Nakano Y.
In Vitro and In Vivo Antiamebic Activity of Iron-Targeting Polypyridine Compounds against Enteric Protozoan Parasite Entamoeba histolytica.
ACS Infectious Diseases
8(3), 457-462 (2022)
doi: 10.1021/acsinfecdis.1c00418
Padhi AK, Kumar A, Haruna KI, et al.
An integrated computational pipeline for designing high-affinity nanobodies with expanded genetic codes.
Briefings in Bioinformatics
22(6), bbab338 (2021)
doi: 10.1093/bib/bbab338
Terasawa K, Kato Y, Ikami Y, et al.
Direct homophilic interaction of LAMP2A with the two-domain architecture revealed by site-directed photo-crosslinks and steric hindrances in mammalian cells.
Autophagy
(2021)
doi: 10.1080/15548627.2021.1911017
Hayashi A, Haruna KI, Sato H, et al.
Incorporation of Halogenated Amino Acids into Antibody Fragments at Multiple Specific Sites Enhances Antigen Binding.
Chembiochem
22(1), 120-123 (2021)
doi: 10.1002/cbic.202000429
Seki E, Yanagisawa T, Kuratani M, et al.
Fully Productive Cell-Free Genetic Code Expansion by Structure-Based Engineering of Methanomethylophilus alvus Pyrrolysyl-tRNA Synthetase.
Acs Synthetic Biology
9(4), 718-732 (2020)
doi: 10.1021/acssynbio.9b00288
Sakamoto K, Hayashi A.
Synthetic tyrosine tRNA molecules with noncanonical secondary structures.
International Journal of Molecular Sciences
20(1), 92 (2019)
doi: 10.3390/ijms20010092
Yanagisawa T, Kuratani M, Seki E, et al.
Structural Basis for Genetic-Code Expansion with Bulky Lysine Derivatives by an Engineered Pyrrolysyl-tRNA Synthetase.
Cell chemical biology
26(7), 936-949 (2019)
doi: 10.1016/j.chembiol.2019.03.008
Teramoto H, Amano Y, Iraha F, et al.
Genetic code expansion of the silkworm Bombyx mori to functionalize silk fiber.
ACS Synthetic Biology
7, 801-806 (2018)
doi: 10.1021/acssynbio.7b00437
Ohtake K, Mukai T, Iraha F, et al.
Engineering an automaturing transglutaminase with enhanced thermostability by genetic code expansion with two codon reassignments.
ACS Synthetic Biology
7, 2170-2176 (2018)
doi: 10.1021/acssynbio.8b00157
Handoko L, Kaczkowski B, Hon C-C, et al.
JQ1 affects BRD2-dependent and independent transcription regulation without disrupting H4-hyperacetylated chromatin states.
Epigenetics
13, 410-431 (2018)
doi: 10.1080/15592294.2018.1469891
Yamaguchi A, Iraha F, Ohtake K, Sakamoto K.
Pyrrolysyl-tRNA Synthetase with a Unique Architecture Enhances the Availability of Lysine Derivatives in Synthetic Genetic Codes.
Molecules (Basel, Switzerland)
23(10), E2460 (2018)
doi: 10.3390/molecules23102460
Members

Team LeaderKensaku Sakamoto
- kensaku.sakamoto[at]riken.jp
(Please replace [at] with @.)
Deputy Team LeaderMotoaki Wakiyama
- motoaki.wakiyama@riken.jp
Senior Research ScientistAkira Wada
- awada[at]riken.jp
Senior Technical ScientistAkiko Matsumoto (Hayash)
- akiko.matsumoto[at]riken.jp
Research ScientistKazumasa Ohtake
- kazumasa.ohtake[at]riken.jp
Technical ScientistMihoko Takahashi
- mihoko.takahashi[at]riken.jp
(Please replace [at] with @)