
Team Leader
Itoshi Nikaido
Ph.D.
Laboratory for Bioinformatics Research
[Affiliation has changed to RIKEN TRIP Headquarters Advanced General Intelligence for Science Program (AGIS) as of April 2025]
E-mailitoshi.nikaido@riken.jp
A multicellular organism is orchestrated by cell growth, death, differentiation, and communication at the single-cell level. To understand various crucial biological phenomena, we should massively perturb and measure transcriptomes and epigenomes at the single-cell level.
Our team will develop novel methods of comprehensive analysis and perturbation of transcriptome and epigenome at the single-cell level, in particular, by applying massively parallel DNA sequencing, genome editing, microfluidics, and machine learning. We focus on the development of methods for quantifying and controlling cell function, fate, and cell-cell communication at the single-cell level.
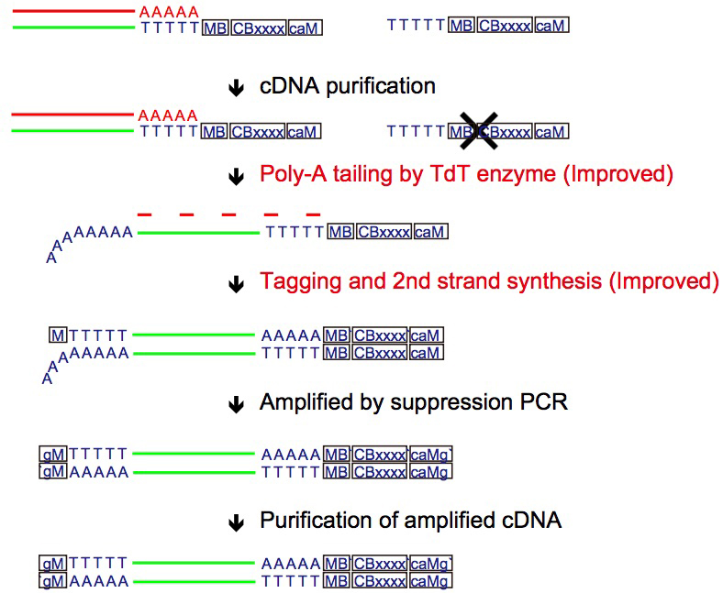
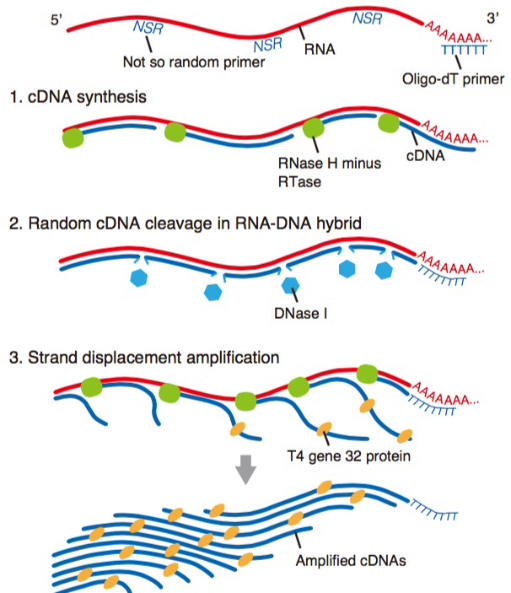
We have already reported novel single-cell RNA-sequencing methods, such as Quartz-Seq and RamDA-seq,which are highly reproducible and sensitive methods of quantifying single-cell transcriptome. Our team will not only develop new techniques but also collaborate with various life scientists within and outside of RIKEN using our new sequencing technologies.
With these techniques, our team seeks to promote the social well-being by contributing insights into how humans can achieve health and longevity.
Research Theme
- Development of novel single-cell omics techniques
- Collaboration with various biologists to apply novel single-cell omics technologies
Selected Publications
Lin CW, Septyaningtrias DE, Chao HW, et al.
A common epigenetic mechanism across different cellular origins underlies systemic immune dysregulation in an idiopathic autism mouse model.
Molecular Psychiatry
27(8), 3343-3354 (2022)
doi: 10.1038/s41380-022-01566-y
Ochiai H, Hayashi T, Umeda M, et al.
Genome-wide kinetic properties of transcriptional bursting in mouse embryonic stem cells.
Science Advances
6(25), eaaz6699 (2020)
doi: 10.1126/sciadv.aaz6699
Ozaki H, Hayashi T, Umeda M, Nikaido I.
Millefy: visualizing cell-to-cell heterogeneity in read coverage of single-cell RNA sequencing datasets.
BMC Genomics
21, 177 (2020)
doi: 10.1186/s12864-020-6542-z
Tsuyuzaki K, Sato H, Sato K, Nikaido I.
Benchmarking principal component analysis for large-scale single-cell RNA-sequencing.
Genome Biology
21, 9 (2020)
doi: 10.1186/s13059-019-1900-3
Mereu E, Lafzi A, Moutinho C, et al.
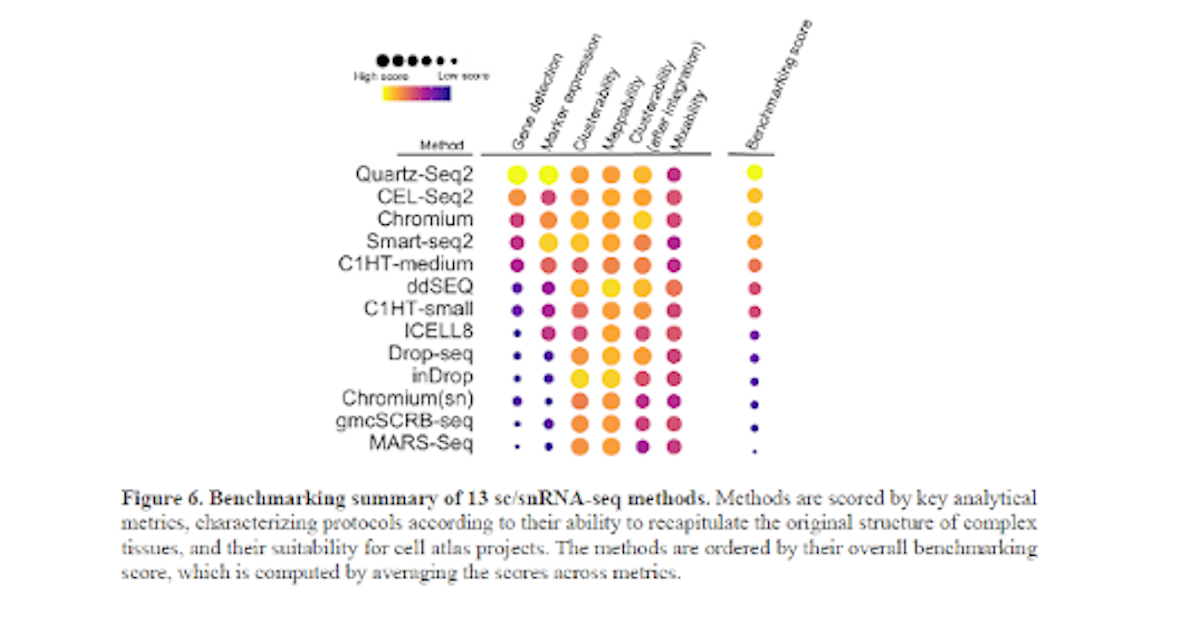
Benchmarking single-cell RNA-sequencing protocols for cell atlas projects.
Nature biotechnology
38(6), 747-755 (2020)
doi: 10.1038/s41587-020-0469-4
Sato K, Tsuyuzaki K, Shimizu K, Nikaido I.
CellFishing.jl: an ultrafast and scalable cell search method for single-cell RNA-sequencing.
Genome Biology
20, 31 (2019)
doi: 10.1186/s13059-019-1639-x
Sasagawa Y, Danno H, Takada H et al.
Quartz-Seq2: a high-throughput single-cell RNA-sequencing method that effectively uses limited sequence reads.
Genome Biology
19, 29 (2018)
doi: 10.1186/s13059-018-1407-3
Hayashi T, Ozaki H, Sasagawa Y, et al.
Single-cell full-length total RNA sequencing uncovers dynamics of recursive splicing and enhancer RNAs.
Nature Communications
9, 619 (2018)
doi: 10.1038/s41467-018-02866-0
Matsumoto H, Kiryu H, Furusawa C, et al.
SCODE: An efficient regulatory network inference algorithm from single-cell RNA-Seq during differentiation.
Bioinformatics
33(15), 2314-2321 (2017)
doi: 10.1093/bioinformatics/btx194
Tsuyuzaki K, Morota G, Ishii M, et al.
MeSH ORA framework: R/Bioconductor packages to support MeSH over-representation analysis.
BMC Bioinformatics
16, 45 (2015)
doi: 10.1186/s12859-015-0453-z
Sasagawa Y, Nikaido I, Hayashi T, et al.
Quartz-Seq: a highly reproducible and sensitive single-cell RNA sequencing method, reveals non-genetic gene-expression heterogeneity.
Genome Biology
14, 3097 (2013)
doi: 10.1186/gb-2013-14-4-r31
Adachi K, Nikaido I, Ohta H, et al.
Context-dependent wiring of Sox2 regulatory networks for self-renewal of embryonic and trophoblast stem cells.
Molecular Cell
52, 380-392 (2013)
doi: 10.1016/j.molcel.2013.09.002
News
Sep. 25, 2020 Research
Scientists identify the molecules responsible for transcriptional bursting

Sep. 1, 2020 BDR News
Dive into BDR's intriguing research
Behind the scenes of cutting-edge research
Apr. 7, 2020 Research
RIKEN group leads world in single-cell transcriptome profiling
May 11, 2018 Research
New single-cell RNA sequencing methods could lead to better regenerative therapies



