Team Director
Ichiro Hiratani
Ph.D.
Laboratory for Developmental Epigenetics
Location Kobe / Developmental Biology Buildings
E-mailichiro.hiratani[at]riken.jp
Please replace [at] with @.
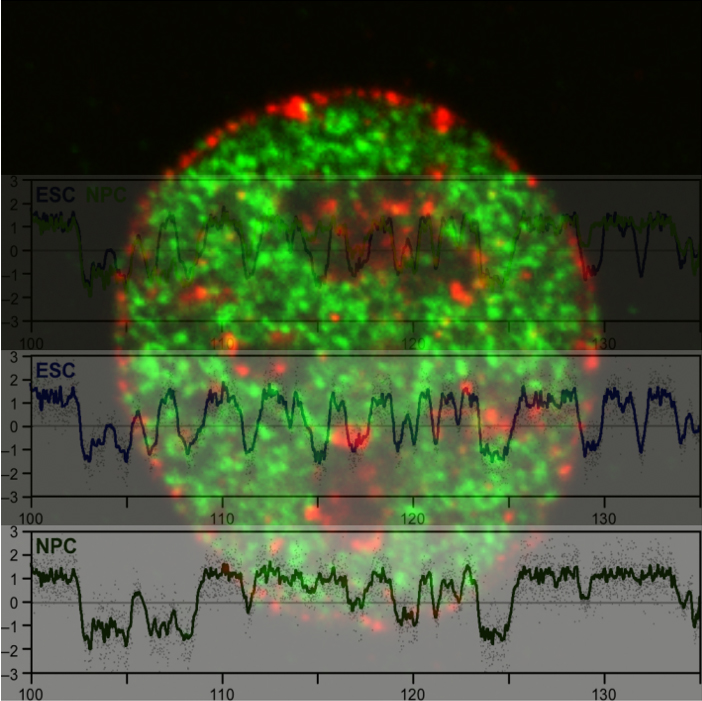
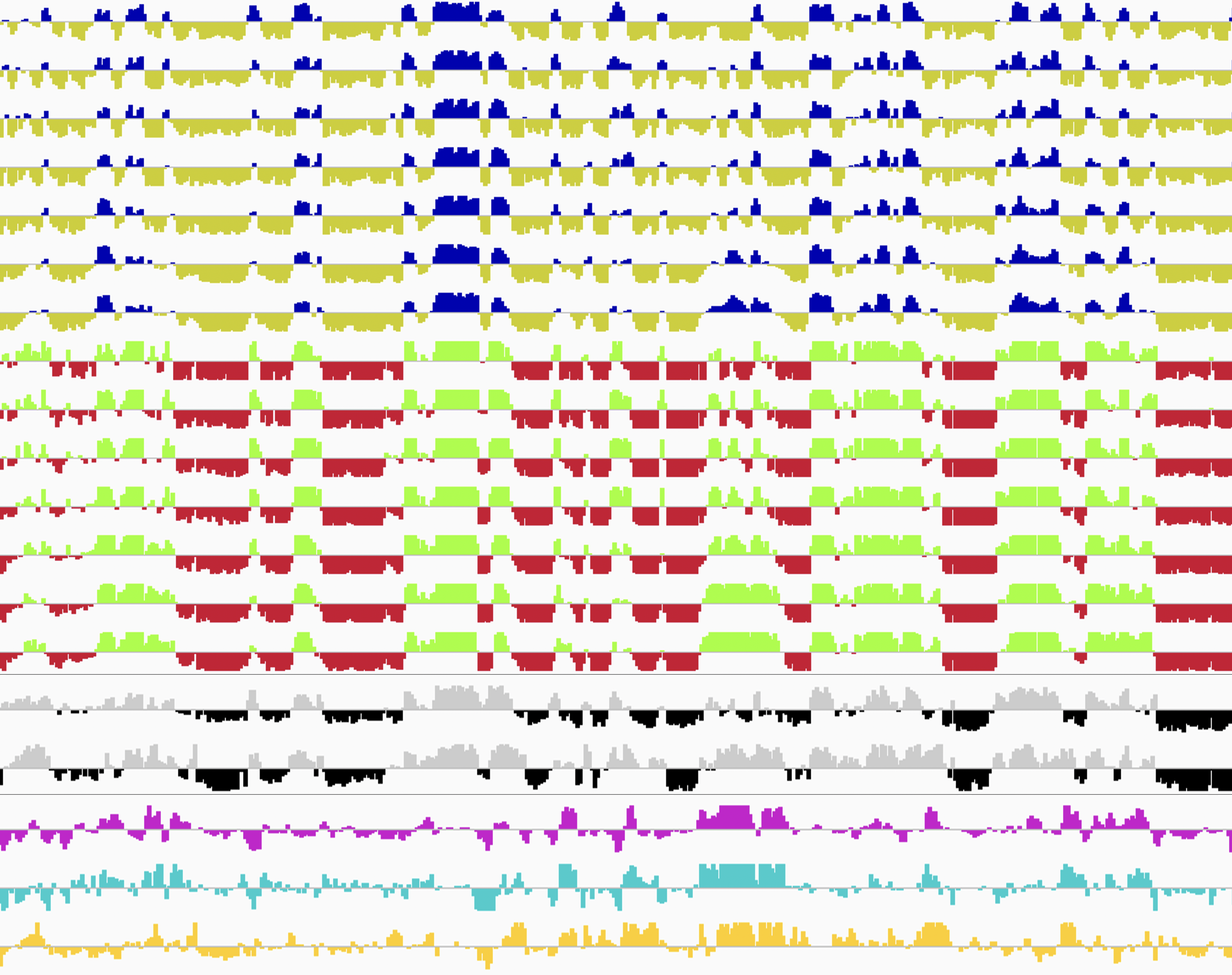
Genomic DNA, which is the source of heredity in all living organisms, is highly compacted in three dimensions (3D) in the cell nucleus. Understanding the regulatory principles of the 3D genome organization is extremely important for life science in general because it would lead to a fundamental understanding of various genome functions such as gene expression. How is the 3D genome organization established and maintained in a cell-type specific manner in various cell types that constitute an organism? How does the 3D genome organization change when cells change their character during development, growth, and differentiation? To answer these questions, our research centers around the regulation of DNA replication, which is known to reflect the 3D genome organization. In particular, we utilize our homemade scRepli-seq method, a single-cell genome-wide DNA replication sequencing technology, and combine it with Hi-C, a genome-wide method to analyze the 3D genome organization. By using a variety of materials from early mouse embryos to cultured cells, we are tackling the aforementioned problems at the single-cell level.
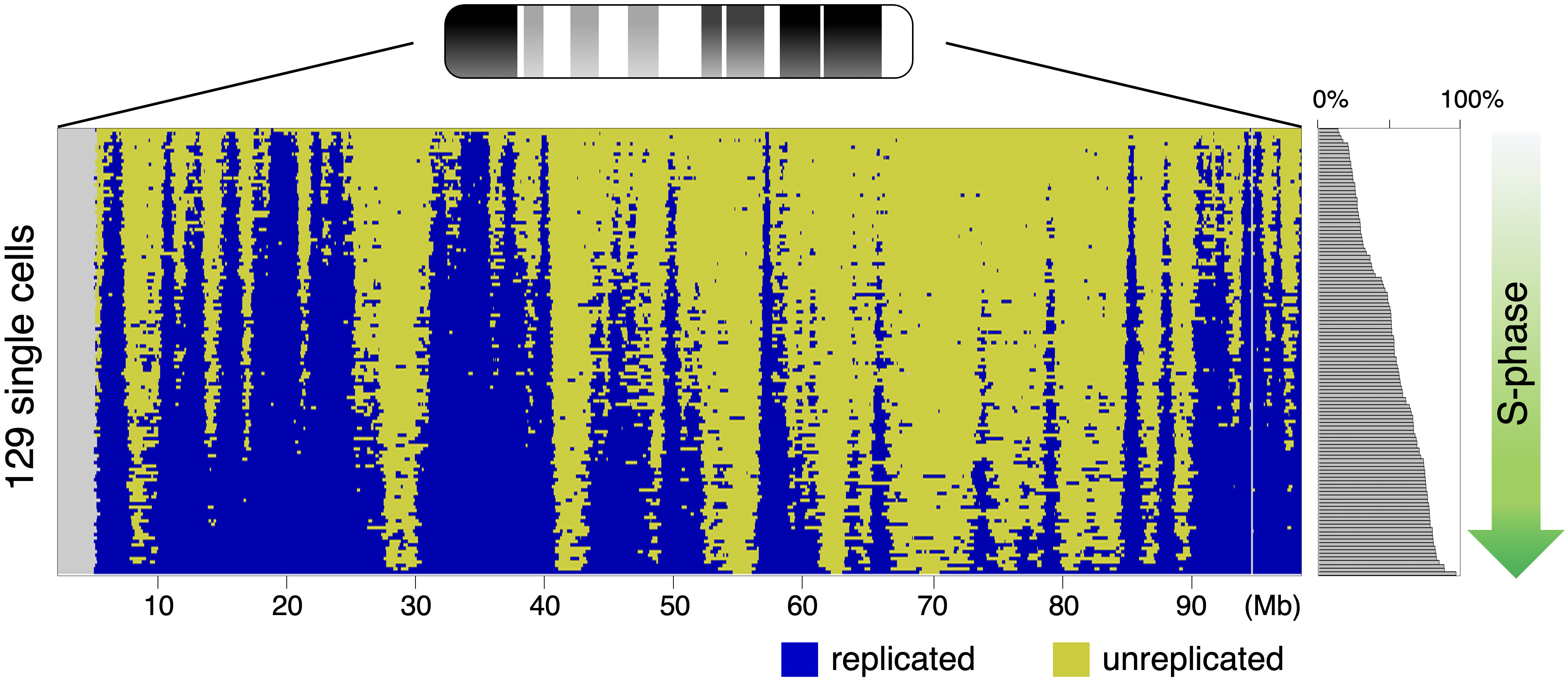
Single-cell genome-wide DNA replication sequencing (scRepli-seq) data
Research Theme
- Developmental dynamics of the 3D genome organization and DNA replication
- Regulatory mechanisms of 3D genome organization
- Development of single-cell genome-wide technologies for studies of the 3D genome organization
Selected Publications
Miura H, Hiratani I.
Cell cycle dynamics and developmental dynamics of the 3D genome: toward linking the two timescales.
Current Opinion in Genetics & Development
73, 101898 (2022)
doi: 10.1016/j.gde.2021.101898
Hada M, Miura H, Tanigawa A, et al.
Highly rigid H3.1/H3.2-H3K9me3 domains set a barrier for cell fate reprogramming in trophoblast stem cells.
Genes & Development
36, 84-102 (2022)
doi: 10.1101/gad.348782.121
Connolly C, Takahashi S, Miura H, et al.
SAF-A promotes origin licensing and replication fork progression to ensure robust DNA replication.
Journal of Cell Science
135(2), jcs258991 (2022)
doi: 10.1242/jcs.258991
Poonperm R, Hiratani I.
Formation of a multi-layered 3-dimensional structure of the heterochromatin compartment during early mammalian development.
Development, Growth & Differentiation
63(1), 5-17 (2021)
doi: 10.1111/dgd.12709
Miura H, Takahashi S, Shibata T, et al.
Mapping replication timing domains genome wide in single mammalian cells with single-cell DNA replication sequencing.
Nature Protocols
15(12), 4058-4100 (2020)
doi: 10.1038/s41596-020-0378-5
Kadota M, Nishimura O, Miura H, et al.
Multifaceted Hi-C benchmarking: what makes a difference in chromosome-scale genome scaffolding?
GigaScience
9(1), giz158 (2020)
doi: 10.1093/gigascience/giz158
Abdalla MOA, Yamamoto T, Maehara K, et al.
The Eleanor ncRNAs activate the topological domain of the ESR1 locus to balance against apoptosis.
Nature Communications
10, 3778 (2019)
doi: 10.1038/s41467-019-11378-4
Miura H, Takahashi S, Poonperm R, et al.
Single-cell DNA replication profiling identifies spatiotemporal developmental dynamics of chromosome organization.
Nature Genetics
51(9), 1356-1368 (2019)
doi: 10.1038/s41588-019-0474-z
Hiratani I, Takahashi S.
DNA Replication Timing Enters the Single-Cell Era.
Genes
10(3), 221 (2019)
doi: 10.3390/genes10030221
Takahashi S, Miura H, Shibata T, et al.
Genome-wide stability of the DNA replication program in single mammalian cells.
Nature Genetics
51(3), 529-540 (2019)
doi: 10.1038/s41588-019-0347-5
Members
Ichiro Hiratani
Team Director
Hisashi Miura
Senior Research Scientist
Rawin Poonperm
Research Scientist
Saori Takahashi
Research Scientist
Asami Oji
Special Postdoctoral Researcher
Akie Tanigawa
Technical Staff I
Takako Ichinose
Technical Staff I
Jothivanan Elumalai
Student Trainee
Linda Jade Choubani
International Program Associate
News
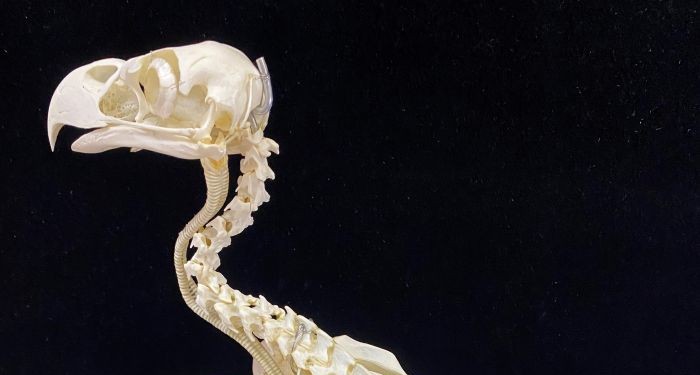
May 1, 2025 Research
Investigating the link between genes and vertebrae in tetrapods

Jan. 23, 2025 BDR News
Team Leader Ichiro Hiratani Awarded Hyogo Prefectural Science Prize
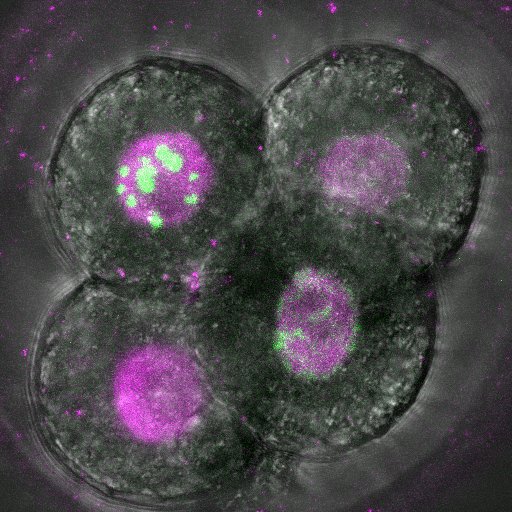
Dec. 24, 2024 Research
DNA copying hits a snag in early embryos
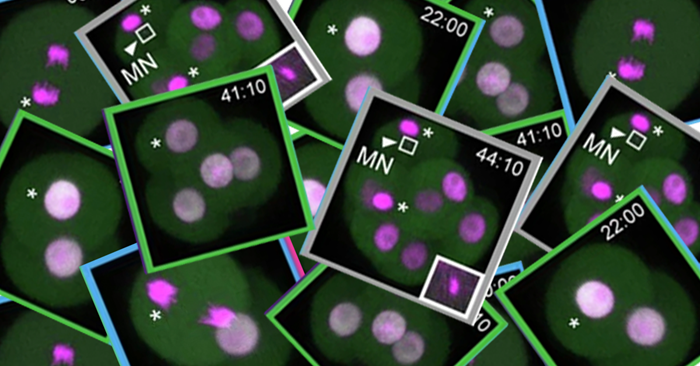
Aug. 31, 2024 Research
Chromosome copy errors pinpointed in embryo development
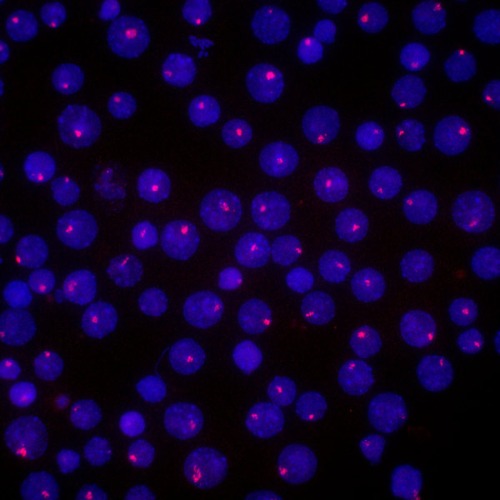
Nov. 13, 2023 Research
Dissecting the structural secrets of the inactive X chromosome
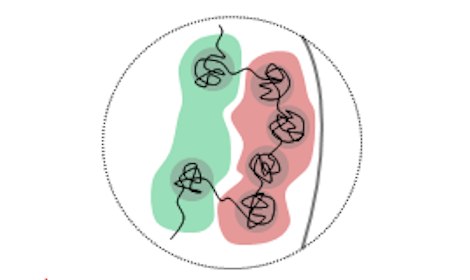
Oct. 9, 2019 Research
How chromosomes change their shape during cell differentiation

Feb. 26, 2019 Research
Scientists lay foundation for single-cell level understanding of DNA replication